Assessment of Glutamine as a Fuel Source for Alveolar Macrophages Exposed to Chronic Ethanol Using an Extracellular Flux Bioanalyzer
概要
Alcohol misuse impairs alveolar macrophage (AM) immunity due to suppressed mitochondrial respiration and bioenergetics. We recently demonstrated that ethanol (EtOH) exposure increases glutamine dependency for mitochondrial respiration in AMs. Herein, methods are provided to determine the usage of glutamine for mitochondrial respiration in EtOH-treated AMs using an extracellular flux bioanalyzer.
Abstract
Alveolar macrophages (AMs) are the first line of cellular defense in the lower airway against pathogens. However, chronic and excessive alcohol use impairs the ability of AMs to phagocytize and clear pathogens from the alveolar space, in part through dysregulated fuel metabolism and bioenergetics. Our prior work has shown that chronic ethanol (EtOH) consumption impairs mitochondrial bioenergetics and increases lactate levels in AMs. Further, we recently demonstrated that EtOH increases glutamine dependency and glutamine-dependent maximal respiration while decreasing flexibility, shifting away from pyruvate-dependent respiration and towards glutamine-dependent respiration. Glutaminolysis is an important compensatory pathway for mitochondrial respiration when pyruvate is used for lactic acid production or when other fuel sources are insufficient. Using a mouse AM cell line, MH-S cells, exposed to either no EtOH or EtOH (0.08%) for 72 h, we determined the dependency of mitochondrial respiration and bioenergetics on glutamine as a fuel source using an extracellular flux bioanalyzer. Real-time measures were done in response to bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl) ethyl sulfide (BPTES), an inhibitor of glutaminase 1, which prevents the enzymatic conversion of glutamine to glutamate, in media vehicle or in response to vehicle alone, followed by testing mitochondrial stress. The step-by-step protocol provided herein describes our methods and calculations for analyzing average levels of glutamine-dependent basal mitochondrial respiration, mitochondrial ATP-linked respiration, maximal mitochondrial respiration, and mitochondrial spare respiratory capacity across multiple biological and experimental replicates.
Introduction
Alcohol use disorder (AUD) affects over 28 million people in the United States1. People with AUD are 2-4 times more likely to develop respiratory infections2,3, increasing risk for premature morbidity and mortality3,4,5. Alcohol misuse increases susceptibility to respiratory infections, in part through suppressing lung immune function. Alveolar macrophages (AMs) are the first line of cellular defense against pathogens in the lower lung, where they phagocytize and clear inhaled microbes. However, chronic alcohol consumption impairs the capacity of AMs to engulf and kill pathogens6,7.
The immune functions of AMs, such as phagocytosis, are energy-demanding processes that require metabolically active pathways necessary for high adenosine triphosphate (ATP) generation. Mitochondrial oxidative phosphorylation is the most efficient cellular process for producing ATP, but chronic ethanol (EtOH) exposure increases mitochondrial-derived oxidative stress, which can result in mitochondrial dysfunction in AMs8,9,10. Mitochondrial respiration measured using cellular oxygen consumption rate (OCR) can be quantified using an extracellular flux bioanalyzer in real-time. Mitochondrial respiration profiles assessed in response to oligomycin (ATP synthase inhibitor), carbonyl cyanide- p-trifluoromethoxyphenylhydrazone (FCCP, proton uncoupler), and rotenone/antimycin A (R/A, mitochondrial complex I and III inhibitors, respectively) are used to determine mitochondrial bioenergetics. Chronic EtOH decreases mitochondrial bioenergetics in AMs, as evidenced by diminished mitochondrial basal respiration, ATP-linked respiration, maximal respiration, and spare respiratory capacity10.
Cells have various levels of dependency on sources of fuels, such as pyruvate, glutamine, or fatty acids, for ATP production. They also have a level of flexibility whereby they can switch fuel usage to regulate cellular bioenergetics. Glutaminolysis is a key pathway for mitochondrial respiration, particularly when pyruvate is shunted towards lactic acid production or when fatty acids are insufficient. Our recent study showed that chronic EtOH exposure increases AM dependency on glutamine and decreases AM flexibility to use glutamine and other fuel sources for mitochondrial respiration11. These results, along with data demonstrating that treatment of EtOH-exposed MH-S cells with the pyruvate oxidation inhibitor UK5099 did not alter mitochondrial bioenergetics compared to media control-exposed MH-S cells treated with UK5099, suggested a shift away from pyruvate-dependent respiration and towards glutamine-dependent respiration in EtOH MH-S cells. However, this shift is insufficient to meet AM bioenergetic demands11. Herein, we describe the methods to assess glutamine as a fuel source for mitochondrial respiration in chronic EtOH-exposed AMs using an extracellular flux bioanalyzer. This protocol was created and optimized for a 96-well (11.04 mm2 area) format and should be adjusted for larger cell culture well sizes. Glutamine dependency for mitochondrial bioenergetics is determined using bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl) ethyl sulfide (BPTES, inhibitor of glutaminase 1) to selectively inhibit the enzymatic conversion of glutamine to glutamate. The data presented in this study are additional biological replicates for the media control- and BPTES-treated experimental groups of untreated control- and chronic EtOH-exposed mouse AM cell line, MH-S cells, that are published in Crotty et al.11.
Protocol
1. Chronic EtOH exposure in vitro
- Culture MH-S cells, a mouse alveolar macrophage cell line, in complete media containing RPMI-1640 with 10% fetal bovine serum, 1% penicillin/streptomycin, 11.9 mM sodium bicarbonate, 40 mg/mL gentamicin, and 50 µM 2-mercaptoethanol at 37°C with 5% CO2 in a humidified incubator.
- Treat cultured MH-S cells with or without EtOH (0.8 mg/mL or 0.08%) for 72 h, where EtOH should be changed every 24 h. Incubate EtOH-treated MH-S cells at 37 °C with 5% CO2 in a separate humidified incubator containing 0.08% EtOH in the incubator water.
2. Plating MH-S cells for assessing glutamine as a fuel source using an extracellular flux bioanalyzer
- Ensure that untreated control (Con)- and chronic EtOH-treated MH-S cells are grown to at least 50% confluency (70%-90% confluency is ideal) in T-75 flasks to passage into a 96-well extracellular flux microculture plate. Prepare a plate map for 5-6 technical replicates per biological replicate of Con and EtOH experimental conditions and media controls for each to compare against BPTES injections (four groups total for biological n= 1).
- Rinse cells with media or phosphate-buffered saline.
- Add 6 mL of serum-containing media to the flask.
- Scrape cells until they are detached and the bottom of the plate is clear. Trypsin is not needed for MH-S cell detachment but may be helpful in other cell types.
- Separate the cells using a serological pipet or 1 mL pipette.
- Transfer the suspended cells to a 15 mL conical tube and centrifuge them at 200 x g for 6 min.
- Aspirate the supernatant.
- Add 1 mL of media and resuspend the cells thoroughly.
- Transfer 10 µL to a microcentrifuge tube and add 10 µL of trypan blue. Mix well.
- Transfer 10 µL of the cell/trypan blue mixture to one side of a hemocytometer or cell counter.
- Count the number of cells using a light microscope or cell counter.
- Calculate the cell density as an average live cell count per milliliter.
- Make calculations for the dilutions that are needed:
Number of wells needed x 10000-15000 cells = total number of cells needed.
Number of wells needed x 80 µL = total volume of resuspended cells needed. - Apply 80 µL of cells to the necessary wells of the 96-well extracellular flux microculture plate.
- Leave the corner wells completely devoid of any cells or media. These wells will be used for background measurements.
- Incubate cells in a 37 °C humidified incubator with 5% CO2 overnight to allow cells to adhere and equilibrate in the extracellular flux microculture plate.
- Treat MH-S cells in the 96-well extracellular flux microculture plate for 72 h using step 1.
- At least 4 h and up to 24 h before running the extracellular flux bioanalyzer experiment, hydrate a 96-well extracellular flux pak cartridge.
- Remove the top portion of the extracellular flux pak and place the probed portion of the flux pak face up on the bench. Apply 200 µL of extracellular flux calibrant solution to each well in the bottom part of the extracellular flux pak cartridge.
- Put the sections together and wrap the extracellular flux pak cartridge in parafilm. Place it in a 37 °C non-CO2 humidified incubator or 37 °C bead bath.
- Turn on the computer containing the assay design and open the running/analysis software attached to the extracellular flux bioanalyzer instrument to allow it to warm up at least 4 h prior to performing the experiment.
3. Using an extracellular flux bioanalyzer to assess glutamine as a fuel source for mitochondrial respiration in MH-S cells
NOTE: The general overview of this procedure is shown in Figure 1.
- Check Con- and EtOH-treated cultured MH-S cells under a light microscope and ensure that cells are healthy and adhered to the wells in a monolayer. Ensure that the cells are at least 70% confluent (90% confluency is ideal) to perform the assay reliably.
- Prepare the extracellular flux base medium with the following supplements:
- Aliquot 35 mL of extracellular flux base medium into a 50 mL tube.
- Add 350 µL of 100 mM sodium pyruvate (1 mM final concentration), 350 µL D-glucose (10 mM final concentration), and 350 µL of GlutaMAX, which is more shelf-stable compared to L-glutamine (2 mM final concentration).
NOTE: Fresh L-glutamine may also be used at the same concentration. - Warm the solution to 37 °C and ensure the pH is 7.4 ± 0.05 at 37 °C.
- Gently aspirate out the growth media from the extracellular microculture plate. Leave as little media as possible without aspirating cells from the bottom. Wash once with a maximum of 50 µL of the extracellular flux-supplemented base medium.
- Add 180 µL of extracellular flux supplemented base medium to each well and incubate the extracellular microculture plate in a 37 °C non-CO2 humidified incubator for 30 min to 1 h to allow for equilibration.
- Dissolve glutamine oxidation test reagents:
- Into the BPTES tube, add 700 µL of extracellular flux-supplemented base medium to make a 120 mM stock solution. Pipette up and down 10 times to properly solubilize the compound.
CAUTION: BPTES causes skin and eye irritation and may cause respiratory irritation. Wear protective gloves, eye protection, and face protection when handling. Dispose of contents and containers to an approved waste disposal facility. - Into the oligomycin tube, add 630 µL of extracellular flux supplemented base medium to make a 100 mM stock solution. Pipette up and down 10 times to properly solubilize the compound.
- Into the FCCP tube, add 720 µL of extracellular flux-supplemented base medium to make a 100 mM stock solution. Pipette up and down 10 times to properly solubilize the compound.
CAUTION: FCCP is harmful if swallowed, causes severe skin burns and eye damage, may cause an allergic skin reaction, and may cause long-lasting harmful effects to aquatic life. Wear protective gloves, protective clothing, eye protection, and face protection when handling. Dispose of contents and containers to an approved waste disposal facility. - Into the R/A tube, add 540 µL of extracellular flux-supplemented base medium to make a 50 mM stock solution. Pipette up and down 10 times to properly solubilize the compound.
CAUTION: Rotenone is fatal if swallowed or if inhaled, causes skin irritation, causes serious eye irritation, may cause respiratory irritation, and is very toxic to aquatic life with long-lasting effects. Wear protective gloves, eye protection, face protection, and respiratory protection. Dispose of contents and containers to an approved waste disposal facility. Antimycin A is fatal if swallowed and is very toxic to aquatic life, with long-lasting effects. Dispose of contents and containers to an approved waste disposal facility.
- Into the BPTES tube, add 700 µL of extracellular flux-supplemented base medium to make a 120 mM stock solution. Pipette up and down 10 times to properly solubilize the compound.
- Prepare working concentrations of substrate oxidation and mitochondrial stress test reagents:
- For BPTES, mix 1,500 µL of extracellular flux supplemented base medium with 500 µL of 120 mM BPTES stock solution to make a 30 µM BPTES working solution.
- For oligomycin, mix 2,520 µL of extracellular flux supplemented base medium with 480 µL of 100 mM oligomycin to make a 16 mM oligomycin working solution.
- For FCCP, mix 2,865 µL of extracellular flux supplemented base medium with 135 µL of 100 mM FCCP to make a 4.5 mM FCCP working solution.
- For R/A, mix 2,700 µL of extracellular flux supplemented base medium with 300 µL of 50 mM R/A to make a 5 mM R/A working solution.
- Load the ports of the upper of the extracellular flux pak cartridge with the following:
- Load the background and media control wells with 20 µL of extracellular flux supplemented base medium into port A, 22 µL of oligomycin into port B, 25 µL of FCCP into port C, and 27 µL of R/A into port D.
- Load the cell-containing wells with 20 µL of media control or BPTES into port A (final concentration of 3 µM for BPTES), 22 µL of oligomycin into port B (final concentration of 2 µM for oligomycin), 25 µL of FCCP into port C (final concentration of 0.5 µM for FCCP), and 27 µL of R/A (final concentration of 0.5 µM for R/A) into port D.
- Perform the extracellular flux bioanalyzer experiment using the computer software program for assay design and analysis that is attached to the instrument using the following timing and injection strategy:
- For the injection with port A for BPTES, set the number of measurements to 6. Set the mix time to 3 min, with a 0-min wait time and a 3-min measurement time.
- For the injection with port B for oligomycin, set the number of measurements to 3. Set the mix time to 3 min, with a 0-min wait time and a 3-min measurement time.
- For the injection with port C for FCCP, set the number of measurements to 3. Set the mix time to 3 min, with a 6-min wait time and a 3-min measurement time.
- For the injection with port D for R/A, set the number of measurements to 3. Set the mix time to 3 min, with a 0-min wait time and a 3-min measurement time.
- Review all set background, media control, and experimental group wells. Save and run the assay. First, load the extracellular flux pak with A-D injections. Calibration will occur until the cell plate is ready to load.
- Remove the cell plate immediately at the end of the assay. Save results to analyze later on another device.
- Check if the cells are still adhered to using a light microscope. Wash 1x with phosphate-buffered saline and lyse cells with a preferred cell lysis buffer. Store at -80 °C or assay immediately for protein concentration.
4. Analysis of results to assess glutamine as a fuel source for mitochondrial respiration in MH-S cells
- Extract protein from cells in each well to normalize oxygen consumption rate (OCR) data to protein:
- Calculate the concentration (nanograms/microliter [ng/µL]) of protein per well.
- Calculate the average protein concentration (ng/µL) for the entire extracellular flux microculture plate.
- Divide each well's protein concentration by the average plate's protein concentration to get a normalization factor.
- Apply the normalization factor to the data file of interest using the computer software program for assay design and analysis.
- Ensure that the OCR values of all background wells are close to 0.
- Deselect any cell wells where OCR is < 10 at baseline. Data will not be deleted, but highlighted wells will show up in the exported data file.
- Export the data from the computer software program for assay design and analysis into a computer spreadsheet program.
- Open the file in the computer spreadsheet program and sort all data by decreasing the values of OCR. Delete background wells, wells that were previously deselected (and had not been deleted), any wells with low cell viability, wells with low cell confluency, wells without full injection strategy completion, wells with inaccurate protein concentration readings, wells with baseline OCR < 10, and any other predetermined criteria.
- Sort all remaining data by measurement, then by group, and then by OCR.
- Average each biological N's technical replicates independently by measurement. Ensure there is a minimum of 18 different values (3 baseline, 3 after media/BPTES, 3 after oligomycin, 3 after FCCP, and 6 after rotenone/antimycin A) per experimental group per biological N after averaging the technical replicates. Do this for each biological N separately so that the standard deviation or standard error of the mean can be calculated later for analyzed data.
- Copy all averaged values into a new spreadsheet, separated by experimental group.
- Average the OCR values by injection strategy in each experimental group.
- Calculate parameters of mitochondrial bioenergetics based on oxygen consumption:
- Calculate basal respiration:
- Calculate basal mitochondrial respiration as follows:
Basal mitochondrial respiration (pmol O2 consumed) = ([AVG OCR measurements #1, 2, 3 – AVG OCR measurements #16, 17, 18] x [Time #3 – Time #1])
NOTE: Basal respiration should be similar between media control and BPTES groups because this is before the inhibitor is injected. - Calculate basal mitochondrial respiration not dependent on glutamine as follows:
Basal mitochondrial respiration not dependent on glutamine (pmol O2 consumed) = ([AVG OCR measurements #4, 5, 6, 7, 8, 9 – AVG OCR measurements #16, 17, 18] x [Time #9 – Time #4])
NOTE: Basal mitochondrial respiration not dependent on glutamine should be less than basal mitochondrial respiration unless cells are not heavily dependent on glutamine. - Calculate basal mitochondrial respiration dependent on glutamine as follows:
Basal mitochondrial respiration dependent on glutamine (pmol O2 consumed) = ([AVG OCR measurements #1, 2, 3 – AVG OCR measurements #4, 5, 6, 7, 8, 9] x [Time #9 – Time #4])
NOTE: Total basal mitochondrial respiration should not be subtracted from basal mitochondrial respiration, and it should not be dependent on glutamine since these are calculated at different time points.
- Calculate basal mitochondrial respiration as follows:
- Calculate ATP-linked respiration as follows:
- Calculate total mitochondrial ATP-linked respiration from all fuels as follows:
Total mitochondrial ATP-linked respiration from all fuels (pmol O2 consumed) = ([AVG OCR measurements #1, 2, 3 – AVG OCR measurements #10, 11, 12] x [Time #12 – Time #10])
NOTE: The average OCR for measurements #10, 11, and 12 should be similar between media control and BPTES groups since oligomycin should strongly inhibit ATP synthase, and inhibiting one fuel pathway should not change the total ATP-linked respiration. - Calculate ATP-linked mitochondrial respiration not dependent on glutamine as follows:
ATP-linked mitochondrial respiration not dependent on glutamine (pmol O2 consumed) = ([AVG OCR measurements #4, 5, 6, 7, 8, 9 – AVG OCR measurements #10, 11, 12] x [Time #12 – Time #10])
NOTE: ATP-linked mitochondrial respiration not dependent on glutamine should be less than ATP-linked mitochondrial respiration unless cells are not heavily dependent on glutamine. - Calculate ATP-linked mitochondrial respiration dependent on glutamine as follows:
ATP-linked mitochondrial respiration dependent on glutamine (pmol O2 consumed) = Total ATP-linked mitochondrial respiration from all fuels – ATP-linked mitochondrial respiration not dependent on glutamine
- Calculate total mitochondrial ATP-linked respiration from all fuels as follows:
- Calculate maximal respiration as follows:
- Calculate total maximal mitochondrial respiration as follows:
Total maximal mitochondrial respiration (pmol O2 consumed) = ([AVG OCR measurements #13, 14, 15 – AVG OCR measurements #16, 17, 18] x [Time #15 – Time #13]) - Calculate maximal mitochondrial respiration not dependent on glutamine as follows:
Maximal mitochondrial respiration not dependent on glutamine (pmol O2 consumed) = ([AVG OCR measurements #13, 14, 15 of BPTES groups – AVG OCR measurements #16, 17, 18 of BPTES groups] x [Time #15 – #Time 13]) - Calculate maximal mitochondrial respiration dependent on glutamine as follows:
Maximal mitochondrial respiration dependent on glutamine (pmol O2 consumed) = Total maximal mitochondrial respiration from all fuels – Maximal mitochondrial respiration not dependent on glutamine
NOTE: This calculation may be used to determine cell dependency for the spare capacity of glutamine compared to other fuels. The greater the maximal mitochondrial respiration, the greater the dependency on glutamine, which can be expressed as BPTES groups – control groups to show a loss in maximal mitochondrial respiration or as a percentage of total respiration.
- Calculate total maximal mitochondrial respiration as follows:
- Calculate spare respiratory capacity as follows:
- Calculate total spare respiratory capacity as follows:
Total spare respiratory capacity (pmol O2 consumed) = ([AVG OCR measurements #13, 14, 15 – AVG OCR measurements #1, 2, 3] x [Time #15 – Time #13]) - Calculate spare respiratory capacity not dependent on glutamine as follows:
Spare respiratory capacity not dependent on glutamine (pmol O2 consumed) = ([AVG OCR measurements #13, 14, 15 of BPTES groups – AVG OCR measurements #1, 2, 3 of BPTES groups] x [Time #15 – Time #13]) - Calculate spare respiratory capacity dependent on glutamine as follows:
Spare respiratory capacity dependent on glutamine (pmol O2 consumed) = Total spare respiratory capacity from all fuels – Spare respiratory capacity not dependent on glutamine
NOTE: This calculation represents the flexibility of cells dependent on glutamine and if they can achieve greater respiration without glutamine when needed.
- Calculate total spare respiratory capacity as follows:
- Calculate basal respiration:
- Subtract media controls by inhibitor groups and normalize pmol O2 consumed relative to the control group if desired.
- Create graphs of OCR bioenergetic profiles (pmol O2/min), basal respiration (pmol O2), ATP-linked respiration (pmol O2), maximal respiration (pmol O2), and spare respiratory capacity (pmol O2) dependent on glutamine.
- Depict mitochondrial bioenergetic endpoint values in bar graphs or express as means of the inhibitor groups relative to the media control groups. If each biological N is calculated separately, then calculate the standard deviation or standard error of the mean for analyzed data across all biological Ns.
5. Statistical analysis
- Perform all statistical analyses using appropriate data analysis software.
- For linear points of OCR bioenergetic profiles, perform two-way ANOVA followed by Tukey's post hoc analyses to compare data from these four experimental groups with two factors (treatment and injection strategy): Con + media, Con + BPTES, EtOH + media, and EtOH + BPTES.
- For bar graphs of basal respiration, ATP-linked respiration, maximal respiration, and spare respiratory capacity dependent on glutamine, conduct Student's t-test analyses to compare data from two experimental groups, Con + BPTES, and EtOH + BPTES.
- Present data as means ± standard error of the mean (SEM). Consider p < 0.05 statistically significant.
Representative Results
Chronic EtOH exposure decreases glutamine dependency in MH-S cells.
Glutaminolysis is a critical pathway for mitochondrial respiration, supporting glutamine as an important fuel source for cellular bioenergetics. To determine whether chronic exposure to EtOH alters the dependency of MH-S cells on glutamine as a fuel source for mitochondrial respiration, MH-S cells were treated with no EtOH control (Con) or EtOH, and OCR was measured over time in response to serial injections of reagents associated with mitochondrial respiration. OCR bioenergetic profiles in response to serial injections of oligomycin (ATP synthase inhibitor), FCCP (proton uncoupler), and R/A (mitochondrial complex I and complex III inhibitors, respectively) are used to calculate other parameters of mitochondrial bioenergetics, such as ATP-linked respiration, maximal respiration, and spare respiratory capacity. All data shown are additional biological replicates of Con + media, Con + BPTES, and EtOH + media, EtOH + BPTES MH-S cell experimental groups that were published in Crotty et al.11. As shown in Figure 2A, oligomycin decreased OCR due to inhibition of complex V, FCCP increased OCR due to mitochondrial uncoupling of membrane potential and mitochondria-dependent ATP generation12, and R/A effectively shut down mitochondria-dependent OCR. These profiles of OCR bioenergetic responses to the serial injections are consistent with the user guide for assessing mitochondrial stress using an extracellular flux bioanalyzer13,14.
After basal OCR was measured in Con- and EtOH-treated MH-S cells, an injection of media control or BPTES, an inhibitor of glutaminase 1, was loaded. Con + media versus Con + BPTES MH-S cells and EtOH + media versus EtOH + BPTES MH-S cells did not demonstrate differences in OCR bioenergetic profiles (Figure 2A). EtOH + BPTES MH-S cells showed a decrease in glutamine-dependent basal respiration compared to Con + BPTES (measurement #4 compared to measurement #3 in Figure 2A and Figure 2B), suggesting that EtOH diminishes MH-S cells' basal glutamine oxidation contributing toward mitochondrial respiration. EtOH + BPTES MH-S cells exhibited a loss of glutamine-dependent-ATP-linked mitochondrial respiration compared to Con + BPTES (Figure 2C). Although EtOH + BPTES MH-S cells demonstrated a blunted loss in glutamine-dependent maximal respiration compared to Con + BPTES (Figure 2D), EtOH + BPTES MH-S cells showed a decrease in glutamine-dependent spare respiratory capacity compared to Con + BPTES (Figure 2E). These results suggest that EtOH MH-S cells have the ability to be more dependent on glutamine for respiration, as when stressed with FCCP, but that they may be unable to do so due to an increase in glutamine demand being outweighed by a decrease in glutamine bioavailability.
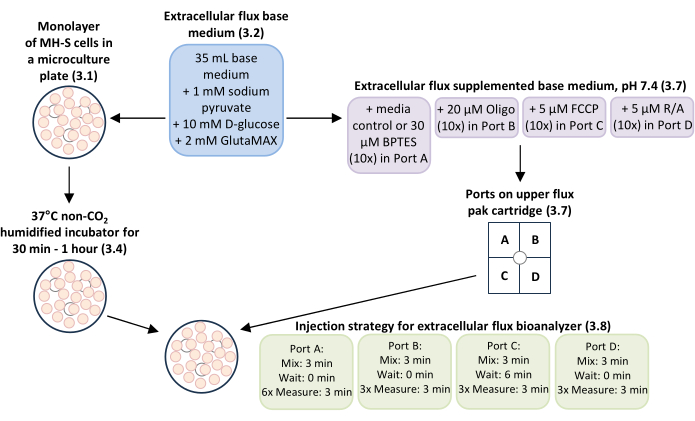
Figure 1: General overview of the workflow for measuring glutamine-dependent mitochondrial respiration and bioenergetics in MH-S cells, a mouse alveolar macrophage cell line. Monolayers of MH-S cells cultured in extracellular flux microculture plate wells (step 3.1) were incubated with extracellular flux base medium with supplements added (step 3.2) at 37°C in a non-CO2 humidified incubator for 30 min-1 h (step 3.4). The extracellular flux base medium with supplements added (step 3.2) was used as media control and to dilute working concentrations of the glutamine oxidation inhibitor BPTES to 3 µM final concentration, the mitochondrial complex V inhibitor oligomycin (Oligo) to 0.5 µM final concentration, the mitochondrial uncoupler carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) to 0.5 µM final concentration, and mitochondrial complex I and complex III inhibitors rotenone/antimycin A (R/A) to 0.5 µM final concentration for loading ports A-D in the upper extracellular flux pak cartridge (step 3.7). The injection strategy for mix, wait, and measure times for the experimental run using the extracellular flux bioanalyzer was: 3 min mix time, 0 min wait time, and 6 measurements of 3 min measure time for Port A (media control or BPTES); 3 min mix time, 0 min wait time, and 3 measurements of 3 min measure time for Port B (Oligo); 3 min mix time, 6 min wait time, and 3 measurements of 3 min measure time for Port C (FCCP); and 3 min mix time, 0 min wait time, and 3 measurements of 3 min measure time for Port D (R/A). Please click here to view a larger version of this figure.
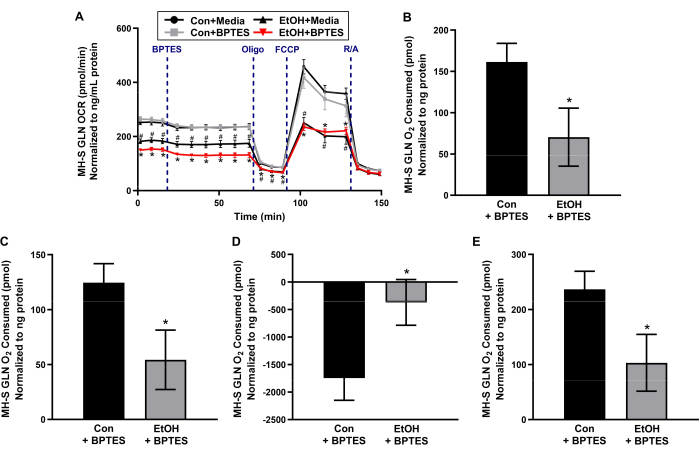
Figure 2: Chronic ethanol (EtOH) decreases glutamine-dependent mitochondrial respiration in MH-S cells, a mouse alveolar macrophage cell line. MH-S cells were untreated (Con) or treated with EtOH (0.08%, 72 h), and oxygen consumption rate (OCR) over time was measured using an extracellular flux bioanalyzer before and after injection with media control or a glutamine (GLN) oxidation inhibitor (3 µM of BPTES), followed by serial injections of mitochondrial complex V inhibitor (0.5 µM of oligomycin, Oligo), mitochondrial uncoupler (0.5 µM of carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, FCCP), and mitochondrial complex I and complex III inhibitors (0.5 µM of rotenone/0.5 µM of antimycin A, R/A). (A) OCR bioenergetic profiles were used to calculate glutamine-dependent (B) basal respiration, (C) ATP-linked respiration, (D) maximal respiration, and (E) spare respiratory capacity. Linear points represent means ± SEM (n = 4, #p < 0.05 versus Con + media and *p < 0.05 versus Con + BPTES, one-way ANOVA with Tukey's post hoc). Bars represent means ± SEM (n = 4, *p < 0.05 versus Con + BPTES, Student's t-test). Data presented are additional biological replicates for the media control- and BPTES-treated experimental groups of untreated control- and chronic EtOH-exposed MH-S cells, which are published in Crotty et al.11. Please click here to view a larger version of this figure.
Discussion
The data presented herein are additional biological replicates for untreated and chronic EtOH-exposed MH-S cells treated with media control or BPTES, which are published in Crotty et al.11. The protocol described is used to assess the dependency of EtOH-exposed MH-S cells on glutamine as a fuel source for mitochondrial respiration and bioenergetics using an extracellular flux bioanalyzer. There are several critical steps in the protocol. Firstly, MH-S cell formation and confluency are important. MH-S cells are initially cultured in extracellular flux microculture wells at low confluency and used when cells are in a monolayer at final confluency of 90%. Since EtOH exposure of MH-S cells was for 72 h, the final confluency of 90% was achieved prior to the experimental run on the extracellular flux bioanalyzer. This is consistent with other reports of cell monolayer formation and 90%-100% confluency before running these experiments15,16,17. Secondly, the normalization of mitochondrial respiration values to protein in each sample is critical. Some studies normalize mitochondrial bioenergetics to cell number18,19,20. However, the serial injection strategy in this assay is meant to sequentially stress mitochondria. In cells that primarily rely on oxidative phosphorylation, such as AMs, this may lead to cell death during the experimental run on the extracellular flux bioanalyzer. Therefore, cell numbers may not be consistent from the beginning of the experiment to the end of the experimental run, and normalization of mitochondrial respiration values to protein is optimal21,22. A protein normalization factor using average protein across all wells is suggested here in order to report more accurate cell-specific OCR profiles. Thirdly, analysis of results across biological replicates is key. At times, representative images of mitochondrial bioenergetic profiles are presented22,23 rather than average mitochondrial respiration across multiple biological replicates10,11,24. However, these representative images do not provide a full picture of the variations between experiments. By calculating each biological replicate separately, the standard error of the mean can show the variation of mean mitochondrial respiration values across all biological replicates and be used to calculate statistical differences between mitochondrial bioenergetic profiles.
Prior to running the assay on the extracellular flux bioanalyzer, the optimal cell density of MH-S cells for culturing in the extracellular flux microculture plate to achieve a monolayer of cells in each well needed to be determined. Since the appropriate concentrations of Oligo and FCCP for MH-S cells also needed to be established, tests of different MH-S cell densities with serial dilutions of Oligo and FCCP were performed21,25. Initial culturing of 10,000-15,000 MH-S cells resulted in a monolayer of cells at 90% confluency after 72 h of exposure with and without EtOH10. Based on these optimization experiments, 2 µM of Oligo and 0.5 µM of FCCP were determined to be the minimum concentrations of these reagents that caused alterations in mitochondrial respiration10.
Real-time measures of mitochondrial respiration at baseline and after treatment with BPTES, an inhibitor of glutaminase 1, which prevents the enzymatic conversion of glutamine to glutamate, is used to determine glutamine dependency for basal respiration in Con- and EtOH-exposed MH-S cells. A limitation of the data presented in this study is that the roles of short and medium-chain fatty acids as sources of fuel for mitochondrial respiration and bioenergetics, in addition to glutamine in EtOH-exposed MH-S cells, have not been investigated. Results from the previous study by Crotty et al.11 suggested that EtOH MH-S cells exhibited a shift from pyruvate-dependent respiration towards glutamine-dependent respiration without full compensation for the loss in total mitochondrial respiration. Further, by performing the same experiment with a combined injection of UK5099 (a pyruvate oxidation inhibitor) and etomoxir (a long-chain fatty acid oxidation inhibitor), the contribution of glutamine in the absence of both pyruvate and fatty acids as fuel sources for mitochondrial respiration could be determined in EtOH-exposed MH-S cells.
Mitochondrial respiration has been measured using a Clark electrode for almost seventy years26, and there are also plate-based fluorescence assays to measure mitochondrial respiration27. However, these methods are significantly more time-intensive and/or lack the sensitivity of measurements that can be achieved with an extracellular flux bioanalyzer. The advantage of using an extracellular flux bioanalyzer to assess glutamine as a fuel source for mitochondrial respiration and bioenergetics is that experiments with multiple biological replicates can be conducted at the same time. Additionally, the extracellular flux bioanalyzer allows for the flexibility to experiment on isolated mitochondria or intact cells. For cells that do not have a high concentration of mitochondria, such as pulmonary artery smooth muscle cells, isolating enough mitochondria from limited samples is challenging. Using the extracellular flux bioanalyzer, mitochondrial respiration can be measured in intact pulmonary artery smooth muscle cells28 or primary mouse AMs10,11. The protocol utilized in this study could be used to assess glutamine as a fuel source for mitochondrial respiration and bioenergetics of other samples of limited sample size, such as primary human AMs. We have previously shown that EtOH-induced mitochondrial-derived oxidative stress and derangements in MH-S cells exhibit a similar profile of mitochondrial dysregulation in primary mouse AMs and human AMs6,7,11,28,29,30. Collectively, studies support using this method of assessing glutamine as a fuel source for mitochondrial respiration and bioenergetics in AMs in pathological conditions that are characterized by mitochondrial-derived oxidative stress.
開示
The authors have nothing to disclose.
Acknowledgements
We acknowledge the contributions of Sarah S. Chang, BS, for the initial cell culture and preparation of MH-S cells for experimentation. This work was supported by NIAAA R01-AA026086 to SMY (ORCID: 0000-0001-9309-0233), NIAAA F31-F31AA029938 to KMC, and NIGMS T32-GM008602 to Randy Hall, Department of Pharmacology and Chemical Biology, Emory University. The contents of this report do not represent the views of the Department of Veterans Affairs or the US Government.
Materials
| 15 mL conical tube | VWR International, Inc. | 21008-670 | Used for preparing stock concentrations of mitochondria-related reagents. |
| 2-mercaptoethanol | Sigma Aldrich Co. | M6250 | Used for culturing MH-S cells. |
| Cell scraper | Dot Scientific, Inc. | 70-1180 | Used for scraping MH-S cells from T-75 flasks. |
| Countess 3 FL Instrument: cell counter | Fisher Scientific Company | AMQAF2000 | Used for counting the number of MH-S cells to plate for experiments. |
| D-glucose | Sigma Aldrich Co. | G8270 | Used for the extracellular flux base medium. |
| Ethanol | Fisher Scientific Company | 4355720 | Experimental treatment for MH-S cells. |
| Fetal bovine serum | Sciencell Research Laboratories | 500 | Used for culturing MH-S cells. |
| Gentamicin | Sigma Aldrich Co. | G1397 | Used for culturing MH-S cells. |
| GlutaMAX | Thermo Fisher Scientific | 35050061 | Used for the extracellular flux base medium. |
| GraphPad Prism 10.2.3 | GraphPad Software | N/A | Software for statistical analysis. Downloadable after purchase at https://www.graphpad.com/features. |
| MH-S cells | American Type Culture Collection | CRL-2019 | Mouse alveolar macrophage cell line. |
| Microcentrifuge tube | USA Scientific, Inc. | 4036-3212 | Used for preparing working concentrations of mitochondria-related reagents. |
| Microsoft 365 Excel: computer spreadsheet program | Microsoft | N/A | Software for data organization and mitochondrial bioenergetics calculations. Downloadable after purchase at https://www.microsoft.com/en-us/microsoft-365/excel. |
| Penicillin/streptomycin | Fisher Scientific Company | 15140122 | Used for culturing MH-S cells. |
| Phosphate buffered saline | VWR International, Inc. | 45000-446 | Used for washing MH-S cells. |
| RPMI-1640 | VWR International, Inc. | 45000-396 | Used for culturing MH-S cells. |
| Seahorse Wave Pro Software: computer software program for assay design and analysis | Agilent Technologies, Inc. | N/A | The computer is attached to the Seahorse XF Pro Analyzer instrument. Downloadable from https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/software-download-for-seahorse-wave-pro-software?productURL=https%3A%2F%2Fwww.agilent.com%2Fen%2Fproduct%2Fcell-analysis%2Freal-time-cell-metabolic-analysis%2Fxf-software%2Fseahorse-wave-pro-software-2007523. |
| Seahorse XF Base Medium: extracellular flux base medium, pH 7.4 | Agilent Technologies, Inc. | 103334-100 | Used for preparing stock and working concentrations of mitochondria-related reagents and culturing MH-S cells prior to experimental runs. |
| Seahorse XF Glutamine Oxidation Stress Test Kit | Agilent Technologies, Inc. | 103674-100 | Contains stock BPTES, oligomycin, FCCP, and rotenone/antimycin A. |
| Seahorse XF Pro Analyzer: extracellular flux bioanalyzer | Agilent Technologies, Inc. | N/A | Extracellular flux bioanalyzer. |
| Seahorse XFe96 FluxPak: 96-well extracellular flux pak cartridges, 96-well extracellular flux microculture plates, and calibrant solution | Agilent Technologies, Inc. | 102416-100 | Used to prepare MH-S cells for assays using an extracellular flux bioanalyzer. |
| Serological pipet | Santa Cruz Biotechnology, Inc. | sc-550678 | Used for removing media from MH-S cells. |
| Sodium bicarbonate | Sigma Aldrich Co. | S6014 | Used for culturing MH-S cells. |
| Sodium pyruvate | Sigma Aldrich Co. | P4562 | Used for the extracellular flux base medium. |
| T-75 flasks | Santa Cruz Biotechnology, Inc. | sc-200263 | Used for culturing MH-S cells. |
参考文献
- . 2021 National Survey on Drug Use and Health (NSDUH). Table 5.6A – Alcohol Use Disorder in Past Year: Among People Aged 12 or Older; by Age Group and Demographic Characteristics, Numbers in Thousands, 2021 Available from: https://www.samhsa.gov/data/sites/default/files/reports/rpt39441/NSDUHDetailedTabs2021/NSDUHDetailedTabs2021/NSDUHDetTabsSect5pe2021.htm (2021)
- Everts, R. J., et al. Nosocomial pneumonia in adult general medical and surgical patients at Christchurch Hospital. N Z Med J. 113 (1111), 221-224 (2000).
- de Roux, A., et al. Impact of alcohol abuse in the etiology and severity of community-acquired pneumonia. Chest. 129 (5), 1219-1225 (2006).
- Moss, M. Epidemiology of sepsis: race, sex, and chronic alcohol abuse. Clin Infect Dis. 41 (Suppl 7), S490-S497 (2005).
- Fernandez-Sola, J., et al. High alcohol intake as a risk and prognostic factor for community-acquired pneumonia. Arch Intern Med. 155 (15), 1649-1654 (1995).
- Yeligar, S. M., Harris, F. L., Hart, C. M., Brown, L. A. Glutathione attenuates ethanol-induced alveolar macrophage oxidative stress and dysfunction by downregulating NADPH oxidases. Am J Physiol Lung Cell Mol Physiol. 306 (5), L429-L441 (2014).
- Yeligar, S. M., Mehta, A. J., Harris, F. L., Brown, L. A., Hart, C. M. Peroxisome proliferator-activated receptor gamma regulates chronic alcohol-induced alveolar macrophage dysfunction. Am J Respir Cell Mol Biol. 55 (1), 35-46 (2016).
- Liang, Y., Harris, F. L., Brown, L. A. Alcohol induced mitochondrial oxidative stress and alveolar macrophage dysfunction. Biomed Res Int. 2014, 371593 (2014).
- Liang, Y., Harris, F. L., Jones, D. P., Brown, L. A. Alcohol induces mitochondrial redox imbalance in alveolar macrophages. Free Radic Biol Med. 65, 1427-1434 (2013).
- Morris, N. L., Harris, F. L., Brown, L. A. S., Yeligar, S. M. Alcohol induces mitochondrial derangements in alveolar macrophages by upregulating NADPH oxidase 4. Alcohol. 90, 27-38 (2021).
- Crotty, K. M., Kabir, S. A., Chang, S. S., Mehta, A. J., Yeligar, S. M. Pioglitazone reverses alcohol-induced alterations in alveolar macrophage mitochondrial phenotype. Alcohol Clin Exp Res (Hoboken). 48 (5), 810-826 (2024).
- Demine, S., Renard, P., Arnould, T. Mitochondrial uncoupling: A key controller of biological processes in physiology and diseases. Cells. 8 (8), 795 (2019).
- Hill, B. G., et al. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem. 393 (12), 1485-1512 (2012).
- Chacko, B. K., et al. The Bioenergetic Health Index: a new concept in mitochondrial translational research. Clin Sci (Lond). 127 (6), 367-373 (2014).
- Fan, Y., et al. Analyzing mitochondrial respiration of human induced pluripotent stem cell-derived myeloid progenitors using Seahorse technology. STAR Protoc. 4 (1), 102073 (2023).
- Gu, X., Ma, Y., Liu, Y., Wan, Q. Measurement of mitochondrial respiration in adherent cells by Seahorse XF96 Cell Mito Stress Test. STAR Protoc. 2 (1), 100245 (2021).
- Plitzko, B., Loesgen, S. Measurement of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in culture cells for assessment of the energy metabolism. Bio Protoc. 8 (10), e2850 (2018).
- Winnica, D., et al. Bioenergetic differences in the airway epithelium of lean versus obese asthmatics are driven by nitric oxide and reflected in circulating platelets. Antioxid Redox Signal. 31 (10), 673-686 (2019).
- Nickens, K. P., Wikstrom, J. D., Shirihai, O. S., Patierno, S. R., Ceryak, S. A bioenergetic profile of non-transformed fibroblasts uncovers a link between death-resistance and enhanced spare respiratory capacity. Mitochondrion. 13 (6), 662-667 (2013).
- Mdaki, K. S., Larsen, T. D., Weaver, L. J., Baack, M. L. Age related bioenergetics profiles in isolated rat cardiomyocytes using extracellular flux analyses. PLoS One. 11 (2), e0149002 (2016).
- Dranka, B. P., et al. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic Biol Med. 51 (9), 1621-1635 (2011).
- Chacko, B. K., et al. Methods for defining distinct bioenergetic profiles in platelets, lymphocytes, monocytes, and neutrophils, and the oxidative burst from human blood. Lab Invest. 93 (6), 690-700 (2013).
- Tyrrell, D. J., Bharadwaj, M. S., Jorgensen, M. J., Register, T. C., Molina, A. J. Blood cell respirometry is associated with skeletal and cardiac muscle bioenergetics: Implications for a minimally invasive biomarker of mitochondrial health. Redox Biol. 10, 65-77 (2016).
- Shah-Simpson, S., Pereira, C. F., Dumoulin, P. C., Caradonna, K. L., Burleigh, B. A. Bioenergetic profiling of Trypanosoma cruzi life stages using Seahorse extracellular flux technology. Mol Biochem Parasitol. 208 (2), 91-95 (2016).
- Nicholls, D. G., et al. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 46, e2511 (2010).
- Chance, B., Williams, G. R. Respiratory enzymes in oxidative phosphorylation. I. Kinetics of oxygen utilization. J Biol Chem. 217 (1), 383-393 (1955).
- Gerencser, A. A., et al. Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal Chem. 81 (16), 6868-6878 (2009).
- Yeligar, S. M., et al. PPARgamma regulates mitochondrial structure and function and human pulmonary artery smooth muscle cell proliferation. Am J Respir Cell Mol Biol. 58 (5), 648-657 (2018).
- Yeligar, S., Tsukamoto, H., Kalra, V. K. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1alpha and microrNA-199. J Immunol. 183 (8), 5232-5243 (2009).
- Yeligar, S. M., Harris, F. L., Brown, L. A. S., Hart, C. M. Pharmacological reversal of post-transcriptional alterations implicated in alcohol-induced alveolar macrophage dysfunction. Alcohol. 106, 30-43 (2023).

.
