Fluorescent End-Labeling and Encapsulation of Long RNAs for Single-Molecule FRET-TIRF Microscopy
概要
This article describes the dual-color labeling of long RNAs at termini positions and their surface immobilization via encapsulation in phospholipid vesicles for single-molecule FRET TIRF microscopy applications. Combining these techniques enables precise visualization and analysis of RNA dynamics at the single-molecule level.
Abstract
Single-molecule Förster Resonance Energy Transfer (smFRET) excels in studying dynamic biomolecules by allowing precise observation of their conformational changes over time. To monitor RNA dynamics with smFRET, we developed a method to covalently label RNAs at their termini with a FRET pair of fluorophores. This direct end-labeling strategy targets the 5'-phosphate by carbodiimide (EDC)/N-hydroxysuccinimide (NHS) activation and the 3'-ribose by periodate oxidation, which can be adapted to other RNAs regardless of their size and sequence to study them independently of artificial modifications. Furthermore, the 5'-EDC/NHS activation is of general interest to all nucleic acids with a 5'-phosphate. The use of commercially available chemicals eliminates the need to synthesize RNA-specific probes.
Total Internal Reflection Fluorescence (TIRF) microscopy requires the surface-immobilized molecules of interest to be within the evanescent field to be illuminated. A sophisticated way of keeping the RNA molecules within the evanescent field is to encapsulate them in phospholipid vesicles. Encapsulation benefits from the best of both worlds, tethering the molecule to the surface while enabling free diffusion of the molecule. We ensure that each vesicle contains only a single RNA molecule, enabling single-molecule imaging. Upon dual-end labeling and encapsulation of the RNA of interest, smFRET measurements offer a dynamic and detailed view of RNA behavior.
Introduction
Förster Resonance Energy Transfer (FRET) is a powerful and sensitive technique for studying inter- and intramolecular interactions of biomolecules at the nanoscale. It is based on the non-radiative energy transfer from an excited donor molecule to a nearby acceptor molecule, which occurs over distances typically between 1 nm and 10 nm. The distance between the donor and acceptor dyes determines the efficiency of this energy transfer, making FRET an invaluable tool for studying molecular dynamics, conformational changes, and interactions in a wide range of biological systems1,2, including RNAs. Total Internal Reflection Fluorescence (TIRF) microscopy has proven to be a powerful technique for smFRET investigations, as it selectively illuminates molecules only near the surface, allowing FRET dynamics of individual molecules with high spatial and temporal resolution. However, before performing smFRET-TIRF experiments, the molecule of interest must first be fluorescently labeled with an appropriate FRET pair and then immobilized on the microscopy surface. The smFRET-TIRF protocol described here was validated using the wild-type group II intron Sc.ai5γ from Saccharomyces cerevisiae mitochondria, flanked by its two exonic sequences (915 nucleotides)3. For a more detailed view of fluorescent labeling group II introns and their immobilization for smFRET TIRF microscopy, refer to our review4.
An ideal site-specific RNA labeling strategy would allow for the precise incorporation of donor and acceptor dyes at predetermined positions without altering the RNA's structure or function, ensuring accurate and efficient FRET measurements. This is challenging due to the chemical similarities among the four nucleobases, which complicates selective labeling. End-labeling attaches donor and acceptor dyes to the RNA ends by targeting the 5'-phosphate and the 3'-ribose. This approach offers a minimally invasive approach while still providing valuable insights into structural dynamics and interactions. Group II intron's ability to self-splice in the presence of Mg2+ restricts the use of metal ion-dependent enzymes. Here, we present an approach to dual end-label long and catalytically active RNAs (ribozymes) that bypass the need for enzymes or the synthesis of specialized probes.
A common approach to tether RNA molecules to the surface for TIRF microscopy is to covalently link a biotin moiety directly to the RNA or to hybridize a biotin-carrying antisense oligonucleotide (ASO)5,6,7. However, this direct immobilization method can introduce artifacts due to RNA-surface interactions, potentially resulting in misfolded RNAs8. An elegant solution to mitigate these immobilization artifacts is to encapsulate RNA in surface-attached nanoscale phospholipid vesicles9,10,11. These vesicles, which are approximately 100 nm in diameter, are anchored to the surface through a biotin-streptavidin linkage12,13,14, allowing the RNA to diffuse freely inside while permitting the exchange of ions across the lipid membrane10. After covalently labeling a large functional RNA3, we present an approach to encapsulate such RNAs in phospholipid vesicles by combining established protocols for surface passivation and vesicle encapsulation, adapted to preserve RNA functionality10,11,14. This dual-end labeling and encapsulation approach achieves a high rate of mono-encapsulation of functional RNAs for smFRET TIRF microscopy.
Protocol
1. RNA dual end-labeling
NOTE: The following protocol describes the site-specific labeling of RNAs with a FRET pair of fluorophores by covalent attachment of a donor dye (sCy3) to the 5'-phosphate and an acceptor dye (Cy5) to the 3'-ribose. A catalytically active long RNA, the group II intron ribozyme, is chosen as the RNA of interest. Table 1 and Figure 1 summarize this dual-end labeling protocol. Perform all steps involving fluorophores under dark conditions.
| Day 0 | ▪ Aliquot 50-75 µg of RNA to a total volume of 55 µL per 1.5 mL tube. | |||||
| Day 1 | 5′-Phosphate activation | |||||
| ▪ Add 45 µL of freshly prepared EDC-NHS, pH 6.0 solution to the RNA in ddH2O to a final volume of 100 μL, mix well and incubate for 4 h at 25 °C and 500 rpm. | ||||||
| ▪ Purification round 1: Overnight EtOH precipitation. | ||||||
| Day 2 | ▪ Precipitate the 5′-activated RNA, wash, and dry. | |||||
| 5′-Dye attachment | ||||||
| ▪ Resuspend in 95 µL of 100 mM MOPS, pH 7.5. | ||||||
| ▪ Add 5 μL of 2 mM amine-functionalized dye solution. | ||||||
| ▪ Mix well and incubate for 16 h at 25 °C and 500 rpm. | ||||||
| Day 3 | ▪ Purification round 2: EtOH precipitation. | |||||
| Day 4 | ▪ Precipitate the 5′-activated RNA, wash, and dry. | |||||
| Blocking step | ||||||
| ▪ Resuspend in 100 μL of 100 mM Tris–HCl, pH 7.5, and incubate for 2 h at 25 °C and 500 rpm. | ||||||
| ▪ Purification round 3: Centrifugal filtration. | ||||||
| → Elute the 5′-labeled RNA. | ||||||
| Day 5 | 3′-Periodate oxidation | |||||
| ▪ Incubate the RNA with 20 mM NaIO4 in 50 mM NaOAc buffer, pH 5.5 in a final volume of 100 μL for 2 h at 25 °C and 500 rpm. | ||||||
| ▪ Quench the excess periodate: Add 30 μL of 50% glycerol, mix well, and incubate for 30 min at 25 °C and 500 rpm. | ||||||
| ▪ Purification round 4: Overnight EtOH precipitation. | ||||||
| Day 6 | ▪ Precipitate the 3′-oxidized RNA, wash, and dry. | |||||
| 3′-Dye attachment | ||||||
| ▪ Resuspend in 95 µL of 50 mM NaOAc, pH 6.0. | ||||||
| ▪ Add 5 μL of 2 mM hydrazide-functionalized dye solution. Mix well and incubate for 16 h at 25 °C and 500 rpm. | ||||||
| Day 7 | ▪ Purification round 5: EtOH precipitation. | |||||
| Day 8 | ▪ Precipitate the labeled RNA, wash, and dry. | |||||
| ▪ Centrifugal filtration. | ||||||
| → Elute the dual-end labeled RNA. | ||||||
Table 1: Protocol summary for RNA dual-end labeling. Please click here to download this Table.
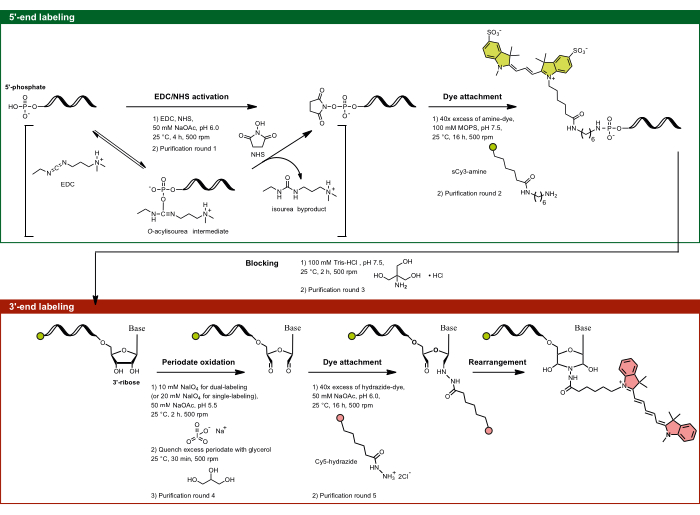
Figure 1: The experimental flow of dual-end labeling by targeting the 5'-phosphate and the 3'-ribose sugar. The 5'-phosphate is activated using EDC in the presence of NHS and is subsequently coupled with the amine-functionalized dye. The 3'-diol moiety of RNA is oxidized by periodate activity to dialdehyde, which further reacts with the hydrazide-functionalized dye. For dual-end labeling, it is important to start with the 5'-labeling to prevent cross-labeling, followed by 3'-labeling with an intermediate blocking step. Abbreviations: EDC = carbodiimide; NHS = N-hydroxysuccinimide; MOPS = 3-morpholinopropane-1-sulfonic acid; NaOAc = sodium acetate. Please click here to view a larger version of this figure.
- 5'-end labeling: Targeting the 5'-phosphate by EDC/NHS activation
NOTE: This 5'-labeling method applies not only to RNA but to any single-stranded nucleic acid containing a 5'-phosphate. The specificity of the reaction is governed by pH dependence, allowing only the 5'-phosphate to be site-specifically targeted, despite the presence of multiple phosphates in the RNA backbone. At pH 6.0, the 5'-phosphate's unique reactivity toward carbodiimides is due to its specific protonation state, where two oxygens are deprotonated, and one remains protonated. This makes the 5'-phosphate reactive, while the fully deprotonated backbone phosphates remain unreactive, enabling selective labeling of the 5'-end via EDC/NHS targeting.- Prepare the EDC-NHS-NaOAc, pH 6.0 solution. Mix 1.5 mg of EDC and 2.0 mg of NHS per aliquot in 35 µL of ddH2O and 10 µL of 0.5 M NaOAc, pH 6.0 (pH adjusted with glacial acetic acid).
- Aliquot the RNA of interest to have approximately 50-75 µg RNA per tube in 55 µL of ddH2O. Add 45 µL of the EDC-NHS-NaOAc, pH 6.0 mixture to the RNA, reaching a total volume of 100 µL with the final concentrations of 78 mM EDC, 174 mM NHS, and 50 mM NaOAc, pH 6.0. Incubate for 4 h at 25 °C while shaking at 500 rpm.
NOTE: Here, 70 µg corresponds to 250 pmol of an in vitro transcribed 915 nt RNA and 2.5 µM in the final reaction volume of 100 µL. - Purification round 1: Purify the 5'-phosphate-activated RNA by EtOH precipitation.
- Prepare a 2 mM sulfonated Cyanine3 amine (sCy3-amine) solution in water.
- Resuspend the activated RNA pellet in 95 µL of 100 mM 3-morpholinopropane-1-sulfonic acid (MOPS) buffer, pH 7.5 (pH adjusted with NaOH). Add 5 µL of 2 mM sCy3-amine solution to the activated RNA. Couple the fluorophore to the activated 5'-phosphate by incubation for 16 h at 25 °C, while shaking at 500 rpm.
NOTE: Store MOPS buffer at 4 °C in the dark. NaOH is chosen for pH adjustment to keep the group II intron ribozyme inactive. The fluorophore should be at least 40-fold excess of the RNA. - Purification round 2: Increase the volume by adding 200 µL of ddH2O to improve the separation. Purify the 5'-phosphate-labeled RNA by EtOH precipitation and repeat until the supernatant is colorless (usually two rounds are needed).
- Blocking step: Resuspend the RNA in 100 µL of 100 mM Tris-HCl, pH 7.5, and incubate for 2 h at 25 °C and 500 rpm. For 5'-end single-labeling only, skip the blocking step, resuspend the RNA in ddH2O, and proceed with step 1.1.8. However, for simplicity, if dual-labeling, separate the 5'end single-labeled control only after step 1.1.9.
NOTE: This step serves to block the activated 5'-phosphates that have not been coupled to an amine-functionalized fluorophore by reacting with a relatively smaller primary amine source (e.g., Tris) to minimize the risk of cross-labeling with the hydrazide-functionalized fluorophore that is used for the 3'-end labeling protocol. - Purification round 3: Remove the free dyes by washing the labeled RNA by centrifugal filtration with a total of at least 10 mL of ddH2O, then elute in ddH2O.
NOTE: The molecular cut-off of the filter should be less than half the size of the nucleic acid. Buffer of choice can be used instead of ddH2O. Elution should follow the manufacturer's instructions, ensuring the sample is not spun to complete dryness. - Determine the RNA and conjugate dye concentrations by UV-Vis spectroscopy.
- 3'-end labeling: Targeting the 3'-ribose by periodate oxidation
- Aliquot the 5'-end labeled RNA in ddH2O to have approximately 50-75 µg of RNA per tube in 90 µL. If the elution volume of the previous step was high, resulting in a low concentration, concentrate the RNA by precipitating and resuspending the pellet in ddH2O. Add 5 µL of 1.0 M NaOAc buffer, pH 5.5 (corresponding to 50 mM NaOAc, pH 5.5 for 100 µL reaction volume).
NOTE: Similar to the 5'-end single-labeled RNA, we routinely prepare 3'-end single-labeled RNA as a control. To do this, aliquot the unlabeled RNA to have approximately 50-75 µg of RNA per tube in 86 µL of ddH2O. - Add 4 µL of freshly prepared 500 mM sodium meta-periodate (NaIO4) stock solution (corresponding to 10 mM NaIO4 for 100 µL reaction volume). For 3'-end single labeling, add 8 µL of NaIO4 stock solution (corresponding to 20 mM NaIO4 for 100 µL reaction volume). Mix thoroughly and incubate for 2 h at 25 °C while shaking at 500 rpm under dark conditions as NaIO4 is light-sensitive.
NOTE: Do not change the order of addition or further increase the incubation time, as this may cause photobleaching of the attached donor dye. The concentration of NaIO4 used is decreased by half for dual-end labeling to minimize the quenching of the already attached dye at the 5'-end. - Stop the reaction by adding 30 µL of 50% glycerol. Incubate for 30 min at 25 °C while shaking at 500 rpm.
NOTE: Glycerol serves as a diol to quench the excess of periodate. - Purification round 4: Add 400 µL of ice-cold EtOH-NaOAc precipitation mixture and perform ethanol precipitation.
- Resuspend the oxidized RNA pellet in 95 µL of 50 mM NaOAc, pH 6.0.
- Hydrazide-dye attachment: Prepare the fluorophore solution by dissolving a few crystals of Cyanine5 hydrazide (Cy5-hydrazide) in DMSO and then diluting with ddH2O to the concentration of 2 mM. If the fluorophore of choice is water-soluble, prepare the solution in ddH2O. Add 5 µL of 2 mM Cy5-hydrazide solution to the oxidized RNA. Mix well and incubate for 16 h at 25 °C while shaking at 500 rpm.
NOTE: The fluorophore should be at least 40-fold excess of the RNA. - Purification round 5: Purify the dual-end-labeled RNA (or 3'-end single labeled RNA) by EtOH precipitation and centrifugal filtration, similar to steps 1.2.6 and 1.2.8, respectively, and elute in ddH2O.
- Calculate the RNA and conjugate dye concentrations by UV-Vis spectroscopy.
- Characterize the labeled RNA using analytical gel-based assays, ensemble fluorescence spectroscopy (see the Representative Results section), and/or analytical HPLC as shown elsewhere3.
- Aliquot the 5'-end labeled RNA in ddH2O to have approximately 50-75 µg of RNA per tube in 90 µL. If the elution volume of the previous step was high, resulting in a low concentration, concentrate the RNA by precipitating and resuspending the pellet in ddH2O. Add 5 µL of 1.0 M NaOAc buffer, pH 5.5 (corresponding to 50 mM NaOAc, pH 5.5 for 100 µL reaction volume).
2. Microfluidic chamber preparation
NOTE: We recommend handling six or eight chambers at a time. Sonication is performed at room temperature unless otherwise indicated. This protocol limits the use of organic solvents, such as acetone, to prevent the solubilization of trace impurities. For alternatives, refer to references15,16.
- Cleaning
- Drill four holes in the quartz slides with a diamond driller according to the scheme given in Figure 2A to form two channels.
NOTE: Although they may break more easily during drilling, glass slides are a cost-effective alternative to quartz slides for objective-based TIRF microscopy setups where the coverslip is the imaging surface (Figure 2B). This is not an option for prism-based TIRF setups, where the microscopy slide is the imaging surface due to background fluorescence.
NOTE: Alternatively, recycle the used microfluidic chambers16. For this, immerse the used chambers overnight in acetone (wrap the Coplin staining jar in aluminum foil to minimize evaporation) in the fume hood, and afterward sonicate for 5 min. Disassemble the chamber, and discard the coverslips and the stickers. If the disassembly does not work immediately, sonicate the chamber in acetone for 10 min more. Proceed with step 2.1.2. - Place the drilled quartz slides and approximately twice as many coverslips (since they may break easily) in a glass Coplin staining jar with a cover. Rinse 3x with ddH2O. Sonicate in ddH2O for 5 min, then rinse 3x with ddH2O.
- Sonicate in 10% laboratory glassware cleaning solution (see Table of Materials) for 30 min at 50 °C. Rinse at least 3x with ddH2O until the detergent bubbles are gone. Sonicate in ddH2O for 5 min. Rinse 3x with ddH2O.
- Sonicate in 1 M KOH solution for 30 min, then leave overnight. Rinse 3x with ddH2O. Sonicate in ddH2O for 5 min. Rinse 3x with ddH2O.
NOTE: Although corrosion due to excessive incubation is a concern previously raised by Chandradoss et al.15, we recommend this long incubation that enables avoiding Piranha etching. - Dry the slides and coverslips with N2(g).
- Treat the dried slides and coverslips with oxygen plasma cleaner for 30 min according to the manufacturer's instructions.
- Drill four holes in the quartz slides with a diamond driller according to the scheme given in Figure 2A to form two channels.
- Aminosilanization
- Prepare the 3% APTES-EtOH solution by thoroughly mixing 288.5 mL of absolute EtOH, 1.5 mL of ddH2O, and 9 mL of (3-aminopropyl)triethoxysilane (APTES) in a 500 mL Erlenmeyer flask.
- Place the clean slides and coverslips in a Coplin staining jar, immerse in the 3% APTES-EtOH solution, sonicate for 1 min, and incubate for 30 min.
- Rinse 3x with absolute EtOH, then 3x with ddH2O.
- Dry the slides and coverslips under N2(g) flow.
- Surface passivation and biotinylation
- Prepare a humid box by filling up an empty pipette tip box up to half with ddH2O. Place the slides inside the box, with the side to be treated facing up.
- Freshly prepare the bPEG-mPEG mixture in a 1.5 mL sterile-clean microcentrifuge tube by gently mixing 2 mg of biotin-polyethylene glycol-succinimidyl valerate 5000 (biotin-PEG-SVA) and 80 mg of methoxy polyethylene glycol-succinimidyl valerate (mPEG-SVA) in 640 µL of 100 mM sodium bicarbonate (NaHCO3) buffer, pH 8.3. Centrifuge the bPEG-mPEG mixture at 16,000 × g for 1 min to remove any air bubbles.
- Carefully remove the supernatant and add a 30 µL droplet to the center of the slide to cover both channels. Place an additional droplet on one side of the slide to have an extra PEGylated coverslip, as they break easily, and having a spare is useful. Finally, cover the drop with a clean coverslip, close the humid box, and perform the PEGylation overnight under dark conditions.
- Thoroughly rinse the PEG-passivated and biotinylated slides and coverslips with ddH2O. Notice the change in the hydrophobicity of the treated surfaces. Dry under N2(g) flow.
- Microfluidic chamber assembly
- Cut a double-sided sticker to create the channels. Attach the sticker to the slide, ensuring it covers the area of interest. Carefully place the coverslip on top, aligning it so that the PEGylated surfaces face each other.
- Place each assembled chamber in a 50 mL centrifuge tube and fill the tubes with N2(g), and store at -20 °C for up to 1 month.
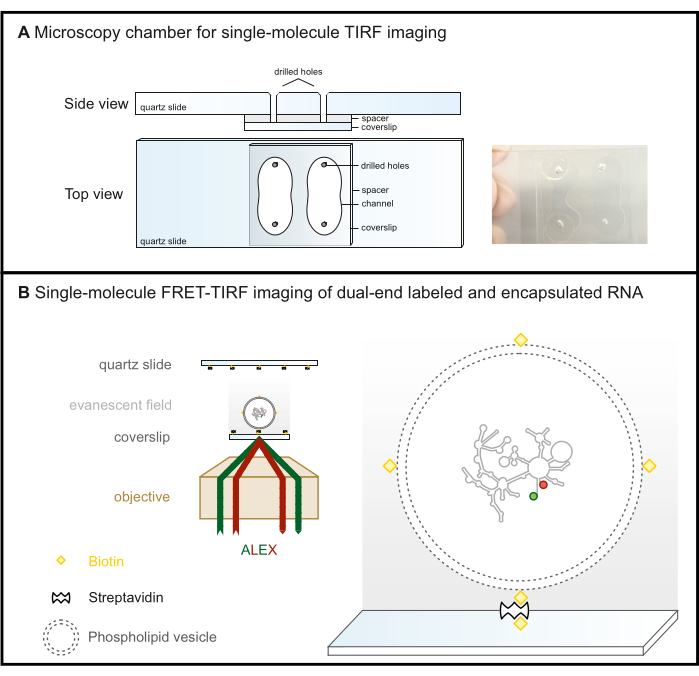
Figure 2: Single-molecule FRET-TIRF microscopy. (A) Microfluidic chamber for TIRF imaging. (B) FRET-labeled RNA is encapsulated in a biotinylated phospholipid vesicle and immobilized on a steptavidin-coated glass surface. This keeps the molecule of interest within the evanescent field (gray gradient), which is created by the incident light that is totally reflected at the critical angle in TIRF microscopy. Here, both fluorophores are subsequently excited with ALEX scheme. Abbreviations: FRET = Förster Resonance Energy Transfer; TIRF = Total Internal Reflection Fluorescence; ALEX = alternative laser excitation. Please click here to view a larger version of this figure.
3. Phospholipid vesicle encapsulation
- Lipid cake preparation
NOTE: Always handle chloroform under a fume hood.- Using a sterile needle, poke holes in the lid (from the inside out) of sterile-clean 2.0 mL microcentrifuge tubes, as shown in Figure 3A.
- Prepare the 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-cap biotinyl (bPE) stock solution by dissolving at least 2 mg of bPE in the appropriate amount of chloroform to a final concentration of 1 mg/mL.
- Prepare the 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) stock solution by dissolving at least 10 mg of DMPC in the appropriate amount of chloroform to a final concentration of 10 mg/mL.
- Prepare the bPE-DMPC mixture (keeping the 1:99 w/w ratio) by mixing 100 µL of bPE and 990 µL of DMPC stock solutions. Distribute 109 µL of the bPE-DMPC mixture per 2 mL tube.
- Place a cardboard cell partition insert of a microtube storage box in a Schlenk flask, serving as a tube holder. Using long tweezers, place the tubes containing the lipid mixture into the tube holder in a 500 mL Schlenk flask (Figure 3B).
NOTE: Make sure that they are upright and not tilted. - Evaporate the chloroform under a low flow of N2(g) overnight (or for at least 2 h until the solvent is completely evaporated).
- Seal the tubes with Parafilm to cover the holes. Store the lipid cakes at -20 °C for up to 1 month.
- RNA encapsulation
- Prepare the following buffers and solutions by adjusting the K+ and Mg2+ concentrations according to the biological system of interest.
- Prepare the 5x standard buffer (5x SB): 2.5 M KCl, 400 mM MOPS; adjust the pH to 6.9 with KOH, sterile filter, and store at 4 °C in the dark (respectively 1x SB: 500 mM KCl, 80 mM MOPS, pH 6.9).
- Prepare the antiblinking buffer (AB): 100 mM MgCl2, Trolox, 1x SB, sterile filter, and store at 4 °C in the dark for up to 1 week.
NOTE: Use a spatula tip of Trolox for 10 mL final volume, vortex to mix, and readjust the pH.
- Assemble the extruder using 100 nm polycarbonate (PC) membrane and 10 mm polyester (PE) drain disc, equilibrate the syringe and the membranes with AB, and heat to 30 °C (or essentially above the DMPC glass transition temperature of 24 °C).
- Mix 5 µL of 1 µM dual-end labeled RNA and 45 µL of AB (to a total volume of 50 µL).
- Hydrate the lipid cake with this RNA solution. At 30 °C, mix for 5 min by shaking at 1,400 rpm, then for 15 min at 700 rpm. Centrifuge for 2 min at 13,000 g and carefully transfer the supernatant into a new tube.
- Dilute the sample with 250 µL of AB.
- Fill the syringe with the RNA-lipid suspension. Extrude 35x at 30 °C on a heating block to encapsulate the dual-end-labeled RNA in phospholipid vesicles of 100 nm diameter.
NOTE: The size distribution of vesicles after using the extruder can be validated by dynamic light scattering (DLS), as shown in reference11.
- Prepare the following buffers and solutions by adjusting the K+ and Mg2+ concentrations according to the biological system of interest.

Figure 3: Lipid cake preparation. (A) The tube caps are poked to enable solvent evaporation. (B) Tubes containing the lipid mixture are placed in a Schlenk flask, and the chloroform is evaporated to obtain a lipid cake. Please click here to view a larger version of this figure.
4. smFRET-TIRF microscopy
- Surface immobilization
- Prepare the following buffers and solutions.
- Prepare the sugar-antiblinking buffer (SAB): 1% D-glucose (w/v) in AB, sterile filter, and store at 4 °C in the dark for up to 1 week.
- Prepare the 100x OSS: 100x Oxygen scavenging system (OSS): 3 mg of glucose oxidase (corresponding to 1.7 U), 10 µL of catalase (corresponding to 22 U) in 90 µL of 1x SB, store at 4 °C in the dark for up to 1 week.
- Freshly prepare the imaging buffer (IB) by mixing 198 µL of SAB and 2 µL of 100x OSS solution.
- Allow the microfluidic chamber to reach room temperature. Flush the chamber 2x with 200 µL of 1x SB.
- Fill the chamber with 50 µL of 20 µL/mL streptavidin solution and incubate for 5 min.
- Flush the chamber with 200 µL of 1x SB and then with 100 µL of AB.
- Immobilize the encapsulated RNA on the surface by adding 75 µL of vesicle suspension and incubate for 10 min.
- Flush the chamber with 200 µL of freshly prepared IB and incubate for 5 min.
- The chamber is now ready for data acquisition (Figure 2B). To prevent evaporation during long measurements, seal the holes as shown in Figure 2A. If leaks from channel to channel are of concern, seal the unused channel before starting the surface immobilization protocol.
NOTE: We recommend performing blank measurements as a quality check for the cleanliness of the microfluidic chambers and phospholipid vesicles against fluorescence contaminations.
- Prepare the following buffers and solutions.
Representative Results
We present the site-specific single- and dual-fluorescent labeling of the 915-nt RNA of interest, the yeast mitochondrial Sc.ai5γ group II intron, flanked by exonic sequences. The FRET fluorophore pair is positioned at the RNA ends via EDC/NHS activation of the 5'-phosphate and periodate oxidation of the 3'-ribose, followed by respective dye attachments. We then verified the RNA-dye conjugates by fluorescent gel electrophoresis, as presented in Figure 4A. The co-migration of RNA and the fluorophores on the agarose gel confirms the successful labeling. Next, as shown in Figure 4B, ensemble fluorescence spectroscopy was used to characterize the dual-labeled group II intron. Energy transfer, that is, FRET, was observed upon excitation of the donor dye proving the dual labeling of the RNA. Notably, consistent with the distance-dependent nature of FRET, the folding of the group II intron RNA in the presence of metal ions led to an increase in FRET efficiency, as evidenced by the decrease in donor emission (green arrow) and a corresponding increase in acceptor (red arrow) emission. This indicates that this FRET labeling tracks the conformational changes of the ribozyme.
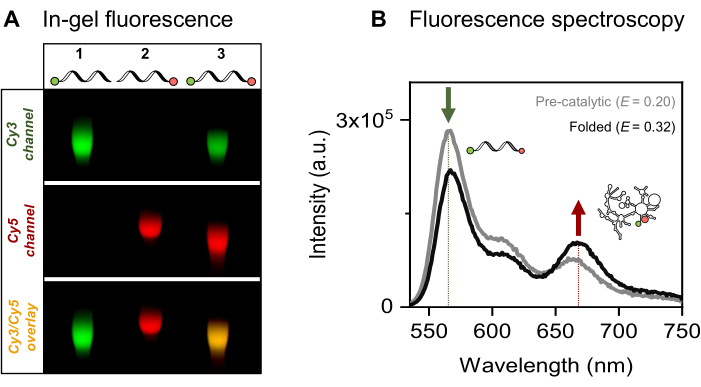
Figure 4: Characterization of the RNA-dye conjugates. (A) Analytical gel-based analysis of single- and dual-fluorescently labeled RNA shows the co-migration of the dyes with the RNA on a 2% agarose gel. Co-localization of the fluorophores in the dual-labeled sample is indicated by the yellow band in the merged image (bottom) of the Cy3 (top, green) and Cy5 (middle, red) channels, visualized under 532 nm and 635 nm illumination, respectively. Lane 1: 5'-sCy3 only labeled RNA, lane 2: 3'-Cy5 only labeled RNA, and lane 3: dual-end (5'-sCy3 and 3'-Cy5) labeled RNA. (B) Ensemble fluorescence spectroscopy confirms dual-labeling. Energy transfer upon donor excitation (λex = 515 nm, λem = 670 nm) verifies that both dyes are successfully coupled to the RNA. The gray curve represents the emission profile of the pre-catalytic RNA, while the black curve demonstrates increased FRET efficiency in the folded group II intron RNA (incubated with 500 mM KCl at 70 °C for 3 min, cooled to 42 °C for 5 min, followed by the addition of 100 mM MgCl2). This figure was adapted from Ahunbay et al.3 Please click here to view a larger version of this figure.
With the fluorescently dual-labeled wild-type group II intron in hand, we are now positioned to explore its dynamics at the single-molecule level. Once encapsulated in phospholipid vesicles, the labeled RNA is immobilized on a microscopy surface at very low surface densities to achieve single-molecule resolution for smFRET-TIRF. As seen in Figure 5A, several individual molecules can be tracked simultaneously. TIRF microscopy enables the real-time monitoring of FRET efficiency and its changes over time. Figure 5B exemplifies the static and dynamic single-molecule FRET traces of the labeled and encapsulated RNA. A typical dynamic trace exhibits anticorrelation between donor and acceptor signals that fluctuate the FRET efficiency. When the acceptor emission upon donor excitation increases, the donor emission correspondingly decreases, indicating a dynamic change in the inter-dye distance. This anticorrelation suggests conformational changes in the RNA molecule.
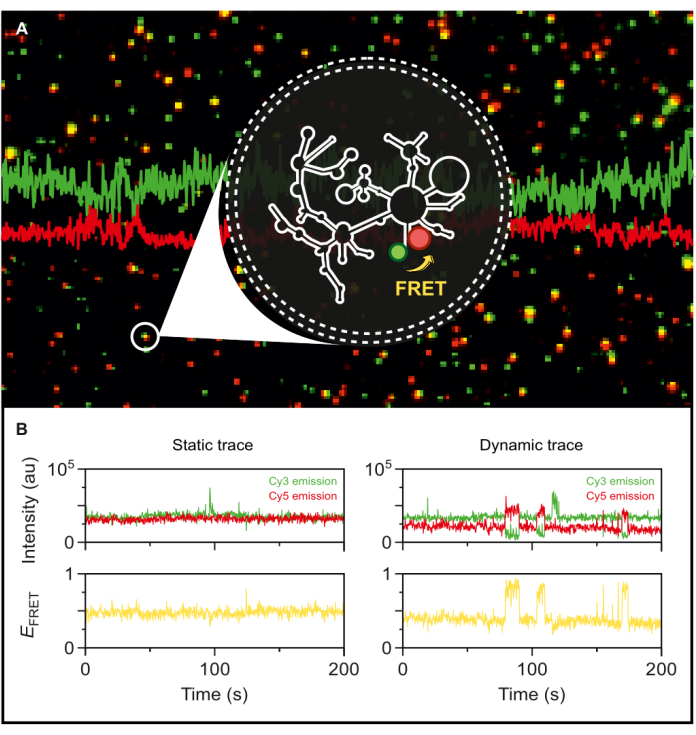
Figure 5: Highly dynamic behavior of group II intron RNA revealed by smFRET. The dual-end labeled group II intron RNA is encapsulated in a phospholipid vesicle and immobilized on the surface for imaging with an objective-based TIRF microscope. (A) Merged image of individual labeled RNA molecules exhibiting donor emission (sCy3, green) and acceptor emission (Cy5, red) upon 532 nm excitation. (B) The typical smFRET trajectories of (left) a static RNA molecule where the donor and acceptor intensities do not fluctuate over time and (right) a dynamic RNA molecule, where the donor and acceptor intensities anticorrelate, with FRET efficiencies in yellow. Single-molecules are localized and analyzed using MASH-FRET17. Direct excitation, bleed-through, and γ-factor corrections are applied. Abbreviations: smFRET = single-molecule Förster Resonance Energy Transfer. Please click here to view a larger version of this figure.
Discussion
FRET at the single-molecule level is unique because it allows the observation and analysis of individual molecules, revealing sample heterogeneity and capturing transient states that can be obscured in ensemble measurements1,2. Observing individual RNA molecules using smFRET provides high-resolution insights into their folding pathways and dynamics. This protocol describes the chemical dual-end labeling of RNA and its surface immobilization via phospholipid vesicle encapsulation, which together enable following dynamic conformational changes via smFRET-TIRF microscopy.
Studying RNA dynamics is a constantly growing field with the need for new site-specific fluorescent labeling strategies. We label the RNA ends by targeting the 5'-phosphate with carbodiimides and the 3'-ribose sugar with periodate. These approaches have been described earlier (5'-end18,19 and 3'-end19,20,21,22) but have not previously been applied to an RNA of similar size to the wild-type Sc.ai5γ group II intron ribozyme, which required optimization. Carbodiimide (e.g., EDC) activation of the 5'-phosphate is reversible. Therefore, imidazole was used to irreversibly react with the O-acylisourea intermediate to form a highly reactive phosphorimidazolide19,20. However, it is now known that at higher pH, carbodiimides can modify nucleobases, specifically guanines and uracils, which has recently led to their use as structural probing agents23,24.
To avoid cross-reactivity, purifying the activated RNA from the EDC prior to raising the pH to 7.5 for the dye coupling step is crucial. However, when we introduced a purification step between the activation and the dye attachment25, we obtained very low yields. Analogously, in protein labeling, surface-accessible lysine residues can be activated with carbodiimides. However, instead of imidazole, which prevents the activation reversal, NHS is routinely used. We adopted this strategy too, thus replacing the phosphorimidazolide intermediate with an NHS-phosphate intermediate. This way we achieved pH control as well as an increased labeling density at lower temperatures and shorter incubation times, i.e., 25 °C for 4 h compared to 37 °C for 16 h. Developed for RNA, this 5'-labeling strategy can be applied to any other single-stranded nucleic acid with a 5'-phosphate.
The 5'-phosphate activation and 3'-ribose oxidation were mutually exclusive, because the chemistries are not orthogonal. To overcome this challenge and avoid cross-labeling, we started with the 5'-end, followed by a blocking step to inhibit the activated but not labeled sites before proceeding with the 3'-end labeling. While oxidizing the 3'-diol, excess sodium meta-periodate (NaIO4) could quench the already attached fluorophore at the 5'-end. Therefore, we reduced the concentration of NaIO4 used for single labeling from 20 mM to 10 mM.
We recommend working with multiple aliquots in parallel instead of scaling up the reactions. This protocol requires multiple ethanol (EtOH) precipitation steps. When working with several aliquots in parallel, prepare a precipitation mixture (30 mL of 100% absolute EtOH and 1 mL of 3 M NaOAc, pH 5.2). NaCl is not used due to its low solubility in EtOH. Precipitate the RNA with 3.1 vol. of this mixture by overnight incubation at -20 °C, followed by centrifugation. Wash the RNA pellet twice with 500 µL ice-cold 70% EtOH, spin down at 4 °C after each time, and dry under vacuum. EtOH precipitation takes advantage of the RNA's insolubility and the free dyes' solubility in 70% ethanol. Centrifugal filtration effectively removes free dyes due to their significant size difference from RNA and facilitates buffer exchange, eliminating salts. In addition to EtOH precipitation and centrifugal filtration methods, free dyes can also be removed using gel extraction and/or chromatography techniques (e.g., HPLC); however, the scale should be adapted accordingly. Do not vortex long RNAs to resuspend, as this may cause mechanical shearing26. The optimal time to pause the protocol is when the RNA is pelleted. We use DNA low-binding tubes to improve nucleic acid recovery. Although the final reaction volume is 100 µL, 1.5 mL (and not lower volume) tubes are preferred for better purification by EtOH precipitation.
Once the unreacted free dyes were removed by precipitation and centrifugal filtration, we confirmed the labeling by fluorescent gel electrophoresis (Figure 4A), UV-Vis spectroscopy, analytical HPLC3. However, it is important to note that these methods cannot distinguish between an RNA molecule carrying both fluorophores and a mixture of RNAs, each labeled with a single color. Similarly, they cannot be used to determine if an RNA molecule carries multiple fluorophores of the same color. Mass spectrometry cannot be used due to size limitations. Ensemble3 and single-molecule FRET spectroscopies corroborate dual-fluorescent labeling, as shown in Figures 4B and 5B. The stoichiometry of 0.5 (sCy3 to Cy5 ratio of 1:1) in smFRET experiments confirms the equal conjugation of the two fluorophores. One concern was the double labeling at the 3'-end by labeling both aldehydes instead of the proposed cyclization. The lack of species with a stoichiometry of 0.25 (sCy3 to Cy5 ratio of 1:2) in smFRET experiments suggests that the dye attachment sterically hinders and prevents the attachment of a second dye.
With this dual-fluorescent labeling, FRET signal changes can be attributed to structural rearrangements throughout RNA folding and catalysis. To maintain fluorescently labeled RNA within the evanescent field for single-molecule TIRF imaging, encapsulation is preferred over direct surface tethering. This approach involves trapping individual RNA molecules within lipid bilayers of vesicles, creating a controlled environment conducive to observing their dynamic behavior. The described protocol enriches mono-encapsulation, as photobleaching in a single step demonstrates11. To understand RNA folding and function, bridging the in vitro and in vivo gap is essential27. Molecular crowding agents can mimic the conditions inside cells to enhance RNA catalysis by group II introns7,28. Alternatively, encapsulation creates constrained microenvironments that promote RNA folding29, bringing our understanding of RNA structure and dynamics closer to a realistic cellular context.
開示
The authors have nothing to disclose.
Acknowledgements
Financial support from the Swiss National Science Foundation [200020_192153 to RKOS], the UZH Forschungskredit [FK-20-081 to EA], the UZH Stiftung für wissenschaftliche Forschung [to RKOS and SZP], Graduate Research Campus (GRC) Short Grant [2024__SG_092 to EA], the Graduate School of Chemical and Molecular Sciences Zurich (CMSZH) and the University of Zurich is gratefully acknowledged.
Materials
| RNA labeling | |||
| 1.5 mL DNA LoBind tubes | Eppendorf | 30108035 | |
| 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC-HCl or EDC) | Thermo Scientific | 11844071 | Pierce EDC, No-Weigh Format. Store at -20 °C |
| 3-morpholinopropane-1-sulfonic acid (MOPS) | Sigma-Aldrich | 69947 | BioUltra, for molecular biology, ≥99.5% (titration) |
| Acetic acid (glacial) | Sigma-Aldrich | 1.00063 | 100%, anhydrous for analysis EMSURE ACS,ISO,Reag. Ph Eur |
| Centrifugal filtration unit | Sartorius | VS0132 | Vivaspin 500, MWCO 50.000, PES, 500 μL |
| Cyanine5 hydrazide (Cy5-hydrazide) | Lumiprobe | 23070 | 5 mg, CAS number 1427705-31-4 |
| Dimethyl sulfoxide (DMSO) | New England Biolabs | 12611P | Molecular biology grade |
| Ethanol | VWR Chemicals | 20821.296P | Absolute ≥99,8% |
| Glycerol | Sigma-Aldrich | G5516 | for molecular biology, ≥99.0% |
| Hydrochloric acid (HCl) | Sigma-Aldrich | 1.00317 | fuming 37%, for analysis EMSURE ACS,ISO,Reag. Ph Eur |
| N-hydroxysuccinimide (NHS) | Sigma-Aldrich | 130672 | 98% |
| Sodium acetate (NaOAc) | Sigma-Aldrich | S8750 | anhydrous, ReagentPlus, ≥99.0% |
| Sodium meta-periodate (NaIO4) | Thermo Fisher Scientific, Life Technologies | 20504 | Pierce product line |
| Sulfo-Cyanine3 amine (sCy3-amine) | Lumiprobe | 213C0 | 5 mg, CAS number 2183440-43-7 |
| Tris(hydroxymethyl)aminomethane (Tris) | Biosolve | 200923 | Molecular biology grade |
| Chamber preparation | |||
| 3-aminopropyl)triethoxysilane (APTES) | Sigma-Aldrich | 440140 | 99% |
| Acetone | Sigma-Aldrich | 1.00014 | for analysis EMSURE ACS,ISO,Reag. Ph Eur |
| Biotin-Polyethylene glycol-Succinimidyl valerate (biotin-PEG-SVA, bPEG) and Methoxy polyethylene glycol-Succinimidyl valerate (mPEG-SVA) | Laysan Bio, Inc. | BIO-PEG-SVA, MW 5,000 and MPEG-SVA, MW 5,000 – Combo Kit | |
| Coverslips | Carl Roth | H876.2 | 24 x 24 mm, 0.13-0.16 mm thickness |
| Deconex 11 universal | Borer Chemie AG | 17416492 | Laboratory glassware cleaning solution |
| Diamond coated core drill bit | Crystalite corporation | 1 mm thickness | |
| Diamond driller | |||
| Ethanol | VWR Chemicals | 20821.296P | Absolute ≥99,8% |
| Glass Coplin staining jar with cover | |||
| Imaging spacer | Grace Bio-Labs | 654008 | SecureSeal, 8 wells, 9 mm × 0.12 mm |
| Oxygen plasma cleaner | Zepto One | Diener | |
| Potassium hydroxide (KOH) | Sigma-Aldrich | P9541 | for molecular biology, ≥99.0% |
| Quartz slides | G. Finkenbeiner, Inc. | 7.5 x 2.5 cm, 1 mm thickness | |
| Sodium bicarbonate (NaHCO3) | Sigma-Aldrich | S6297 | BioXtra, 99.5-100.5% |
| Lipid cake preparation | |||
| 16:0 Biotinyl Cap PE, bPE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (sodium salt), C53H98N4O11PNaS) | Avanti Polar Lipids | 870277P | Stable for 1 year at -20 °C |
| 14:0 PC, DMPC (1,2-ditetradecanoyl-sn-glycero-3-phosphocholine or 1,2-dimyristoyl-sn-glycero-3-phosphocholine, C36H72NO8P) | Avanti Polar Lipids | 850345P | |
| 2.0 mL microcentrifuge tubes, Safe-Lock | Eppendorf | 0030120094 | Autoclave to sterilize |
| 500 mL Schlenk flask | |||
| Chloroform | Merck | 1.02445 | for analysis EMSURE ACS, ISO, Reag. Ph Eur |
| Parafilm | |||
| Surface immobilization via encapsulation | |||
| (±)-6-Hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox®) | Thermo Fischer | 53188-07-1 | 97% |
| 3-morpholinopropane-1-sulfonic acid (MOPS) | Sigma-Aldrich | 69947 | BioUltra, for molecular biology, ≥99.5% (titration). Store at +4 °C in the dark. |
| Adhesive seal tabs | Grace Bio-Labs | 629200 | |
| Catalase | Sigma-Aldrich | 9001-05-2, C30 | from bovine liver, aqueous suspension, 10,000-40,000 units/mg protein |
| D-glucose | Sigma-Aldrich | G7528 | ≥99.5% (GC), BioXtra |
| Extruder polycarbonate (PC) membrane | Avanti Polar Lipids | 610005-1EA | 0.1 μm, 19 mm |
| Extruder set with heating block | Avanti Polar Lipids | 610000 | |
| Glucose oxidase | Sigma-Aldrich | 9001-37-0 | from Aspergillus niger, Type VII, lyophilized powder, ≥100,000 units/g solid (without added oxygen) |
| Magnesium chloride (MgCl2) | Sigma-Aldrich | 7786-30-3, M1028 | for molecular biology, 1.00 M ± 0.01 M solution |
| Polyester (PE) drain disc, membrane | Whatman, Cytiva | 230300 | 10 mm |
| Potassium chloride (KCl) | Sigma-Aldrich | 60128 | BioUltra, for molecular biology, ≥99.5% (AT) |
| Potassium hydroxide (KOH) | Sigma-Aldrich | P9541 | for molecular biology, ≥99.0% |
| Streptavidin | Thermo Fischer | 434301 |
参考文献
- Eitan, L., et al. FRET-based dynamic structural biology: Challenges, perspectives and an appeal for open-science practices. Elife. 10, e60416 (2021).
- Taekjip, H., et al. Fluorescence resonance energy transfer at the single-molecule level. Nat Rev Methods Primers. 4, 22 (2024).
- Ahunbay, E., Steffen, F. D., Zelger-Paulus, S., Sigel, R. K. O. Chemical dual end-labeling of large ribozymes. Methods Mol Biol. 2439, 191-204 (2022).
- Ahunbay, E., Zelger-Paulus, S., Sigel, R. K. O. Group II Introns: Highly structured yet dynamic. Chimia (Aarau). 77 (4), 235-241 (2023).
- Steiner, M., Karunatilaka, K. S., Sigel, R. K. O., Rueda, D. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci USA. 105 (37), 13853-13858 (2008).
- Karunatilaka, K. S., Solem, A., Pyle, A. M., Rueda, D. Single-molecule analysis of Mss116-mediated group II intron folding. Nature. 467 (7318), 935-939 (2010).
- Paudel, B. P., Fiorini, E., Börner, R., Sigel, R. K. O., Rueda, D. S. Optimal molecular crowding accelerates group II intron folding and maximizes catalysis. Proc Natl Acad Sci USA. 115 (47), 11917-11922 (2018).
- Saha, R., Verbanic, S., Chen, I. A. Lipid vesicles chaperone an encapsulated RNA aptamer. Nat Commun. 9 (1), 2313 (2018).
- Boukobza, E., Sonnenfeld, A., Haran, G. Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. J Phys Chem B. 105 (48), 12165-12170 (2001).
- Cisse, I., Okumus, B., Joo, C., Ha, T. Fueling protein DNA interactions inside porous nanocontainers. Proc Natl Acad Sci USA. 104 (31), 12646-12650 (2007).
- Zelger-Paulus, S., Hadzic, M. C. A. S., Sigel, R. K. O., Börner, R. Encapsulation of fluorescently labeled RNAs into surface-tethered vesicles for single-molecule FRET studies in TIRF microscopy. Methods Mol Biol. 2331, 1-16 (2020).
- Okumus, B., Wilson, T. J., Lilley, D. M. J., Ha, T. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys J. 87 (4), 2798-2806 (2004).
- Ishitsuka, Y., Okumus, B., Arslan, S., Chen, K. H., Ha, T. Temperature-independent porous nanocontainers for single-molecule fluorescence studies. Anal Chem. 82 (23), 9694-9701 (2010).
- Liu, B., Mazouchi, A., Gradinaru, C. C. Trapping single molecules in liposomes: surface interactions and freeze-thaw effects. J Phys Chem B. 114 (46), 15191-15198 (2010).
- Chandradoss, S. D., et al. Surface passivation for single-molecule protein studies. J Vis Exp. (86), e50549 (2014).
- Paul, T., Myong, S. Protocol for generation and regeneration of PEG-passivated slides for single-molecule measurements. STAR Protoc. 3 (1), 101152 (2022).
- Hadzic, M. C. A. S., Kowerko, D., Börner, R., Zelger-Paulus, S., Sigel, R. K. O., Farkas, D. L., Nicolau, D. V., Leif, R. C. Detailed analysis of complex single molecule FRET data with the software MASH. Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues IX. SPIE 971119, (2016).
- Chu, B. C., Wahl, G. M. Le Orgel. Derivatization of unprotected polynucleotides. Nucleic Acids Res. 11 (18), 6513-6529 (1983).
- Qin, P. Z., Pyle, A. M. Site-specific labeling of RNA with fluorophores and other structural probes. Methods. 18 (1), 60-70 (1999).
- Whitfeld, P. R. A method for the determination of nucleotide sequence in polyribonucleotides. Biochem J. 58 (3), 390-396 (1954).
- Zamecnik, P. C., Stephenson, M. L., Scott, J. F. Partial purification of soluble RNA. Proc Natl Acad Sci USA. 46 (6), 811-822 (1960).
- Proudnikov, D., Mirzabekov, A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 24 (22), 4535-4542 (1996).
- Wang, P. Y., Sexton, A. N., Culligan, W. J., Simon, M. D. Carbodiimide reagents for the chemical probing of RNA structure in cells. RNA. 25 (1), 135-146 (2019).
- Mitchell, D., et al. In vivo RNA structural probing of uracil and guanine base-pairing by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). RNA. 25 (1), 147-157 (2019).
- Rinaldi, A. J., Suddala, K. C., Walter, N. G. Native purification and labeling of RNA for single molecule fluorescence studies. Methods Mol Biol. 1240, 63-95 (2015).
- Liu, T., Patel, S., Pyle, A. M. Making RNA: Using T7 RNA polymerase to produce high yields of RNA from DNA templates. Methods in Enzymology. , (2023).
- Leamy, K. A., Assmann, S. M., Mathews, D. H., Bevilacqua, P. C. Bridging the gap between in vitro and in vivo RNA folding. Q Rev Biophys. 49, e10 (2016).
- Fiorini, E., Börner, R., Sigel, R. K. O. Mimicking the in vivo Environment–The Effect of Crowding on RNA and Biomacromolecular Folding and Activity. Chimia. 69 (4), 207-212 (2015).
- Peng, H., Lelievre, A., Landenfeld, K., Müller, S., Chen, I. A. Vesicle encapsulation stabilizes intermolecular association and structure formation of functional RNA and DNA. Curr Biol. 32 (1), 86-96.e6 (2022).

.
