Oral Biofilm Analysis of Palatal Expanders by Fluorescence In-Situ Hybridization and Confocal Laser Scanning Microscopy
概要
We present a protocol for structural and compositional analysis of natural oral biofilm from orthodontic appliances with in situ hybridization (FISH) and confocal laser scanning microscopy (CLSM). Oral biofilm samples were collected from palatal expanders, scraping acrylic-resin flakes off their surface and referring them for molecular processing.
Abstract
Confocal laser scanning microscopy (CLSM) of natural heterogeneous biofilm is today facilitated by a comprehensive range of staining techniques, one of them being fluorescence in situ hybridization (FISH).1,2 We performed a pilot study in which oral biofilm samples collected from fixed orthodontic appliances (palatal expanders) were stained by FISH, the objective being to assess the three-dimensional organization of natural biofilm and plaque accumulation.3,4 FISH creates an opportunity to stain cells in their native biofilm environment by the use of fluorescently labeled 16S rRNA-targeting probes.4-7,19 Compared to alternative techniques like immunofluorescent labeling, this is an inexpensive, precise and straightforward labeling technique to investigate different bacterial groups in mixed biofilm consortia.18,20 General probes were used that bind to Eubacteria (EUB338 + EUB338II + EUB338III; hereafter EUBmix),8-10 Firmicutes (LGC354 A-C; hereafter LGCmix),9,10 and Bacteroidetes (Bac303).11 In addition, specific probes binding to Streptococcus mutans (MUT590)12,13 and Porphyromonas gingivalis (POGI)13,14 were used. The extreme hardness of the surface materials involved (stainless steel and acrylic resin) compelled us to find new ways of preparing the biofilm. As these surface materials could not be readily cut with a cryotome, various sampling methods were explored to obtain intact oral biofilm. The most workable of these approaches is presented in this communication. Small flakes of the biofilm-carrying acrylic resin were scraped off with a sterile scalpel, taking care not to damage the biofilm structure. Forceps were used to collect biofilm from the steel surfaces. Once collected, the samples were fixed and placed directly on polysine coated glass slides. FISH was performed directly on these slides with the probes mentioned above. Various FISH protocols were combined and modified to create a new protocol that was easy to handle.5,10,14,15 Subsequently the samples were analyzed by confocal laser scanning microscopy. Well-known configurations3,4,16,17 could be visualized, including mushroom-style formations and clusters of coccoid bacteria pervaded by channels. In addition, the bacterial composition of these typical biofilm structures were analyzed and 2D and 3D images created.
Protocol
1. Specimen collection
- Remove fixed palatal expanders after 4 months of intraoral service. Use NOLA Dry Field System to isolate the appliances before removing them (Figures 1 and 2).
- Use sterilized pliers, gloves and trays to remove expanders without adding contamination (Figures 3 and 4).
- Store objects in Sarstedt 120 ml vials at -20°C. Take to laboratory on ice and process within 24 hours.
2. Biofilm fixation
- Scrape off biofilm flakes with a sterile scalpel, or collect pieces with sterile forceps.
- Add ice-cold 4% PFA solution until sample is well covered.
- Incubate the mixture at +4°C (do not freeze) for 3 to 12 hours. Longer fixation times or higher fixation temperatures may render the cell envelopes of gram-negative cells less permeable to oligonucleotide probes.
- Remove PFA solution and wash with ice-cold 1X PBS. Repeat this step 2-3 times to remove residual PFA.
- Resuspend the sample in 1 vol. ice-cold 1X PBS and add 1 vol. ice-cold 96% (v/v) ethanol.
- Store the sample at -20°C. Samples fixed according to this protocol can be stored for several months to years.
3. Dehydration of fixed Samples
- Apply 5-30 μl of PFA-fixed sample material to a microscope slide.
- Dry at 46°C for about 15 min or at room temperature for longer. Thaw the lysozyme and the formamide.
- Add 250 μl of lysozyme (1mg/ml) at room temperature for 10 min, thus facilitating penetration of the probes into the cells by partial destruction of the cell walls.
- Dip slide into 50%, 80% and 96% (v/v) ethanol for 3 min each. The dehydrating effect of the ethanol concentration series will disintegrate the cell membranes.
- Dry the slides at 46°C for 10 min.
4. In-situ hybridization
- Prepare 1 ml of fresh hybridization buffer (see Table 1 for concentrations). Formamide concentrations used in this study: 10% (EUBmix), 20% (Bac303, POGI, MUT590) or 45% (LGCmix).
- Thaw the oligonucleotide probe solutions. Thawed probes should be kept on ice and protected from light.
- Add 2 μl of each probe to 200 μl of hybridization buffer, mix well and apply the mixture to the dehydrated sample on a microscope slide.
- Place a piece of tissue paper into a square Petri dish and pour the remaining hybridization buffer onto the tissue paper.
- Immediately place the slide horizontally into the dish and close the dish. Incubate in an oven at 46°C for 1-5 hours (90 min will suffice in most cases). The dish functions as a moisture chamber preventing evaporation of hybridization solution from the slide. In particular, evaporation of formamide can cause non-specific probe binding to non-target cells.
- Prepare 50 ml of washing buffer (see Table 2 for concentrations). Prepare the washing buffer in a 50 ml tube and preheat to 48°C in a water bath. The washing step is performed at 48°C.
- Remove the dish with the slide from the hybridization oven. Immediately wash off the hybridization buffer with a small volume of pre-warmed washing buffer, and transfer the slide into the remaining washing buffer.
- Place the tube containing the washing buffer and the slide back into the water bath and incubate at 48°C for 10-15 min.
- Remove slide from the tube and dip into ice-cold ddH2O for 2-3 seconds to eliminate residual washing buffer.
- Air-dry the slide as quickly as possible (the use of compressed air is recommended). Fast drying will reduce probe dissociation.
- Dried slides can be stored in the dark at -20°C for several weeks without significant loss of probe-conferred fluorescence signal.
5. Microscopy
- After FISH and washing, apply two drops of antifadent close to the left and right ends of a slide (frozen slices should be warmed to room temperature prior to this step).
- Place a microscope cover slip on top and wait until the antifadent has spread over the entire slide. Note that an excessive amount of antifadent can blur the microscope image.
- Observe the samples under a confocal laser scanning microscope equipped with suitable filters or lasers. We used a Leica TCS unit (HCX PL APO/63x; NA 1.2). Data can be analyzed with software such as IMARIS or AMIRA.
- Slides embedded in antifadent can be stored at +4°C (do not freeze) for up to 7 days before the probe-conferred fluorescence begins to decline. Alternatively, the antifadent can be removed with ddH2O, and the dried slides can be stored at -20°C for an extended period of time.
6. Representative Results:
Scraping biofilm off fixed orthodontic appliances (Figure 5) yields suitable flakes (Figure 6) that can be hybridized directly onto coated glass slides for microscopy. In this way, different groups of orobiome bacteria can be identified in their natural three-dimensional environment by tagging bacterial rRNA with differently labeled specific probes (Figures 7 and 8). In Figure 7, biofilm was stained with EUBmix (green, all bacteria) and LGCmix (yellow, Firmicutes). Firmicutes appear in green, as they were stained with yellow and blue. In Figure 8, biofilm was stained with EUBmix (red, all bacteria), Bac303 (blue, Bacteroidetes) and POGI (yellow, Porphyromonas gingivalis). Porphyromonas gingivalis is shown in yellowish white, as all three probes bind to its DNA and the overlap of colors results in a white signal. Morphological differences between groups of bacteria can also be identified (Figures 9 and 10). Large clusters of coccoid bacteria are shown in Figure 9, where staining was performed with EUBmix (green, all bacteria). Different shapes of oral bacteria were visualized in Figure 10, where coccoid and filamentous bacteria can be distinguished by staining with EUBmix (red). Also, a typical mushroom-like structure of the biofilm can be processed via 3D modeling of the CLSM data (Figures 11 and 12) Click here to watch a movie of the 3D modeling.
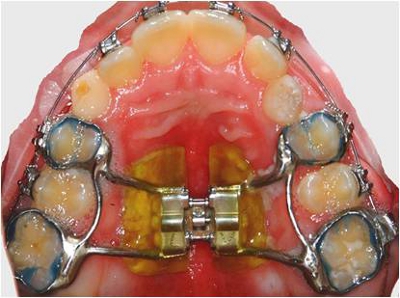
Figure 1. Fixed palatal expander in situ.
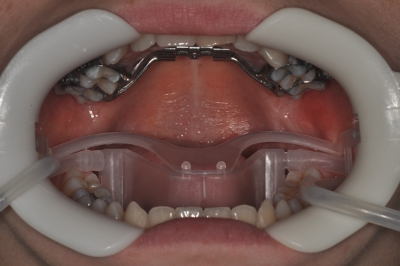
Figure 2. Nola Dry Field System.

Figure 3. Sterilized pliers, gloves and tray.
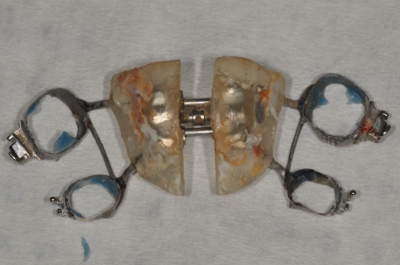
Figure 4. Removed expander.
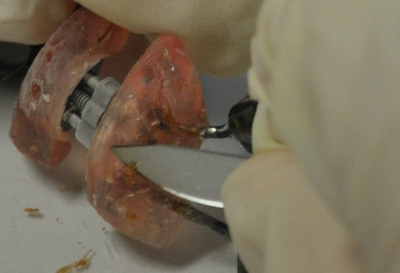
Figure 5. Scraping off biofilm with a sterile scalpel.

Figure 6. Resin flakes directly hybridized onto coated glass slides.
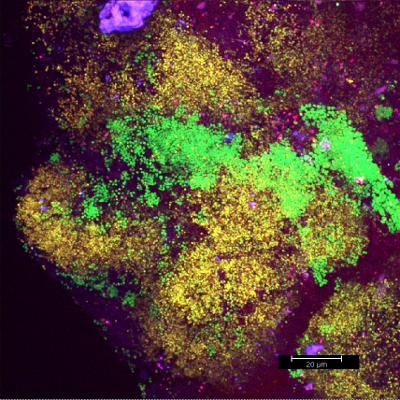
Figure 7. CLSM image: differentiation of a specific bacterial group. 2D overlay of 3D CLSM stack data. Biofilm stained with EUBmix (green, all bacteria) and LGCmix (yellow, Firmicutes).
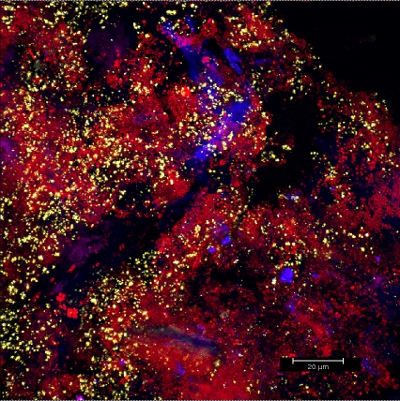
Figure 8. CLSM image: differentiation of a specific bacterial group. 2D overlay of 3D CLSM stack data. Biofilm stained with EUBmix (red, all bacteria), Bac303 (blue, Bacteroidetes) and POGI (yellow, Porphyromonas gingivalis).
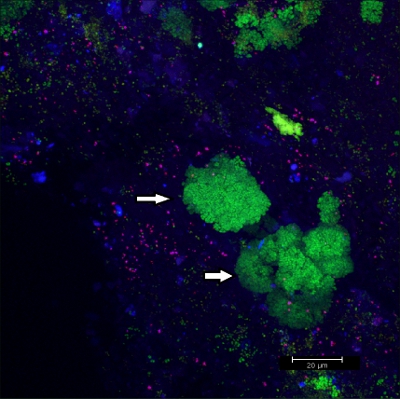
Figure 9. CLSM image: differentiation of specific morphologies. 2D overlay of 3D CLSM stack data. Biofilm stained with EUBmix (green, all bacteria). Large clusters of coccoid bacteria (arrows).
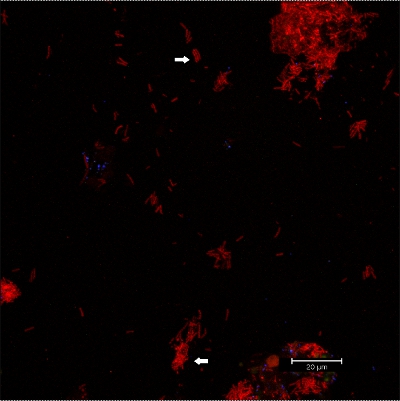
Figure 10. CLSM images: differentiation of specific morphologies. 2D overlay of 3D CLSM stack data. Biofilm stained with EUBmix (red, all bacteria). Coccoid (arrow below) and filamentous bacteria (arrow above) can be distinguished.
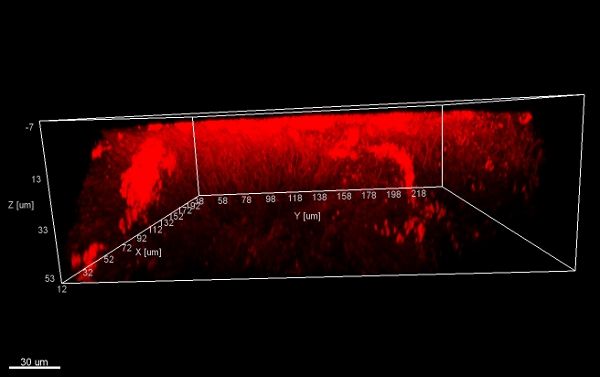
Figure 11. Mushroom structure, 3D views from below. Stacks of biofilm flakes (scraped off the surface of an orthodontic appliance), stained with EUBmix and processed with IMARIS (CLSM image).

Figure 12. Mushroom structure, 3D side view. Stacks of biofilm flakes (scraped off the surface of an orthodontic appliance) processed with IMARIS (CLSM image).
| Hybridization buffer (200 μl) | |||
| Formamide concentration | 10% | 20% | 45% |
| 5 M NaCl | 36 | 36 | 36 |
| 1 M Tris-HCl | 4 | 4 | 4 |
| 2% SDS | 1 | 1 | 1 |
| FA | 20 | 40 | 90 |
| ddH2O | 138 | 118 | 68 |
| Any FISH probe | 2 | 2 | 2 |
Table 1. Constituents of hybridization buffer (concentrations in μl).
| Washing buffer (50 ml) | |||
| Formamide concentration | 10% | 20% | 45% |
| 5 M NaCl | 4500 | 2150 | 300 |
| 1 M Tris-HCl | 1000 | 1000 | 1000 |
| 0.5 M EDTA | 0 | 500 | 500 |
| ddH2O | 44 500 | 46 350 | 48 200 |
Table 2. Constituents of washing buffer (concentrations in μl).
Movie 1. Click here to watch the movie.
Discussion
The protocol herein described is a highly workable approach to staining biofilms collected from hard materials. Sampling and hybridization are the most critical steps. During sampling, care must be taken that slices of adequate thickness are collected to ensure that the biofilm structure remains intact. During hybridization, it is essential to avoid excessive fluctuations in temperature, thus avoiding non-specific binding, or loss of binding, of the fluorescently labeled probes. This FISH protocol is an elegant method to stain normal heterogeneous biofilm directly on glass slides without disrupting its composition. The slides can immediately be used for microscopy without a need to move the sample once again. In this way, an improvement over staining in tubes is achieved by less shear force being applied to the biofilm. Slices > 50 μm thick can be prepared and investigated in this fashion.
開示
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Hygiene Fonds of the Medical University Graz. All subjects gave their informed consent. Institutional approval of the study protocol was obtained from the Medical University Graz.
Materials
| Oligonucleotides | Target organism | Fluorochrome |
|---|---|---|
| EUB338 – 338III | Eubacteria | Cy3 |
| LGC354 A-C | Firmicutes | FITC |
| Bac303 | Bacteroidetes | Cy5 |
| Mut590 | Streptococcus mutans | FITC |
| POGI | Porphyromonas gingivalis | FITC |
Table 3. Oligonucleotides (refer to probeBase13 for details).
| Component | Company | コメント |
|---|---|---|
| 4% paraformaldehyde | 4% solution in PBS | |
| 1 x PBS | 3 x solution prepared | |
| ddH2O | Millipore | |
| Ethanol (100%) | Merck | |
| Lysozyme | Roth | 100 mg/ml stock solution |
| Formamide | Roth | 99.5% solution |
| 5 M NaCl | Roth | |
| 1 M Tris-HCl | Roth | |
| 2% SDS | Roth | |
| FISH probes (oligonucleotides) | Invitrogen, Sigma | |
| ProLong Gold antifadent | Invitrogen | |
| Nail polish | ||
| Equipment | ||
| Incubator | Thermo Scientific | |
| Water bath | IKA Tez | |
| 50 ml tubes | Sarstedt | |
| Polysine-coated slides | Thermo Scientific |
Table 4. Materials and equipment.
参考文献
- Sunde, P. T. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. 微生物学. 149, 1095-1102 (2003).
- Sussman, M., Loya, Y., Fine, M., Rosenberg, E. The marine fireworm Hermodice carunculata is a winter reservoir and spring-summer vector for the coral-bleaching pathogen Vibrio shiloi. Environ. Microbiol. 5, 250-255 (2003).
- Kolenbrander, P. E. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66, 486-505 (2002).
- Zijnge, V. Oral biofilm architecture on natural teeth. PLoS One. 5, e9321-e9321 (2010).
- Thurnheer, T., Gmur, R., Giertsen, E., Guggenheim, B. Automated fluorescent in situ hybridization for the specific detection and quantification of oral streptococci in dental plaque. J. Microbiol. Methods. 44, 39-47 (2001).
- Hojo, K., Nagaoka, S., Ohshima, T., Maeda, N. Bacterial interactions in dental biofilm development. J. Dent. Res. 88, 982-990 (2009).
- Jung, D. J. Visualization of initial bacterial colonization on dentine and enamel in situ. J. Microbiol. Methods. 81, 166-174 (2010).
- Al-Ahmad, A. Bacterial colonization of enamel in situ investigated using fluorescence in situ hybridization. J. Med. Microbiol. 58, 1359-1366 (2009).
- Bryers, J. D. Medical biofilms. Biotechnol. Bioeng. 100, 1-18 (2008).
- Cardinale, M., Vieira de Castro, J., Muller, H., Berg, G., Grube, M. In situ analysis of the bacterial community associated with the reindeer lichen Cladonia arbuscula reveals predominance of Alphaproteobacteria. FEMS Microbiol. Ecol. 66, 63-71 (2008).
- Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., Dewhirst, F. E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721-5732 (2005).
- Chin, M. Y., Busscher, H. J., Evans, R., Noar, J., Pratten, J. Early biofilm formation and the effects of antimicrobial agents on orthodontic bonding materials in a parallel plate flow chamber. Eur. J. Orthod. 28, 1-7 (2006).
- Loy, A., Maixner, F., Wagner, M., Horn, M. probeBase–an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic. Acids. Res. 35, (2007).
- Diaz, P. I. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 72, 2837-2848 (2006).
- Dige, I., Nilsson, H., Kilian, M., Nyvad, B. In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning microscopy and fluorescence in situ hybridization. Eur. J. Oral Sci. 115, 459-467 (2007).
- Kolenbrander, P. E. Bacterial interactions and successions during plaque development. Periodontology. 42, 47-79 (2000).
- Kolenbrander, P. E., Palmer, R. J., Periasamy, S., Jakubovics, N. S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 8, 471-480 (2010).
- Chalmers, N. I. Use of quantum dot luminescent probes to achieve single-cell resolution of human oral bacteria in biofilms. Appl. Environ. Microbiol. 73, 630-636 (2007).
- Wecke, J. A novel technique for monitoring the development of bacterial biofilms in human periodontal pockets. FEMS Microbiol. Lett. 191, 95-101 (2000).
- Moter, A., Gobel, U. B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods. 41, 85-112 (2000).

