Immunostaining-Based Detection of Dynamic Alterations in Red Blood Cell Proteins
概要
Capturing dynamic changes in the protein activation of enucleated red blood cells poses methodological challenges, like the preservation of dynamic changes to acute stimuli for later assessment. The presented protocol describes sample preparation and staining techniques that enable preservation and analysis of relevant protein changes and subsequent detection.
Abstract
Antibody labeling of red blood cell (RBC) proteins is a commonly used, semi-quantitative method to detect changes in overall protein content or acute alterations in protein activation states. It facilitates the assessment of RBC treatments, characterization of differences in certain disease states, and description of cellular coherencies. The detection of acutely altered protein activation (e.g., through mechanotransduction) requires adequate sample preparation to preserve otherwise temporary protein modifications. The basic principle includes immobilizing the target binding sites of the desired RBC proteins to enable the initial binding of specific primary antibodies. The sample is further processed to guarantee optimal conditions for the binding of the secondary antibody to the corresponding primary antibody. The selection of non-fluorescent secondary antibodies requires additional treatment, including biotin-avidin coupling and the application of 3,3-diaminobenzidine-tetrahydrochloride (DAB) to develop the staining, which needs to be controlled in real-time under a microscope in order to stop the oxidation, and thus staining intensity, on time. For staining intensity detection, images are taken using a standard light microscope. In a modification of this protocol, a fluorescein-conjugated secondary antibody can be applied instead, which has the advantage that no further development step is necessary. This procedure, however, requires a fluorescence objective attached to a microscope for staining detection. Given the semi-quantitative nature of these methods, it is imperative to provide several control stains to account for non-specific antibody reactions and background signals. Here, we present both staining protocols and the corresponding analytical processes to compare and discuss the respective results and advantages of the different staining techniques.
Introduction
Red blood cells (RBCs) traverse the cardiovascular system for 70 to 140 days, with a mean RBC age of approximately 115 days1,2. Senescent or damaged RBCs are removed from the circulation by erythrophagocytosis, an efficient clearing process driven by macrophages3. The predetermined lifespan of these cells is one consequence of surrendering the cell organelles, including the nucleus, mitochondria, and ribosomes, during differentiation and maturation4. Thus, circulating RBCs are devoid of a translational machinery, precluding the synthesis of new proteins3. It follows that dynamic, post-translational modifications to existing proteins represent the only viable mechanism of acute, biochemical regulation in response to extracellular and intracellular stressors acting on RBCs5.
Mechanical forces appear to be chief extracellular cues that cause the activation or modulation of biochemical pathways within RBCs. The discovery of the mechanosensitive protein, Piezo1, in RBC membranes6 inspired several lines of research investigating mechanically-activated signaling in these cells7. For example, recent advances have shown that the physical properties of RBCs are actively regulated by acute and dynamic changes of proteins8, which includes post-translational phosphorylation and ubiquitination9. Since these normal modifications differ in certain diseases9,10,11, it seems to be of scientific and clinical interest to determine the activation state of RBC proteins, specifically in relation to mechanobiological processes.
The determination of acute changes in RBC protein activation states poses some methodological challenges. For instance, the storage of RBC samples for later analysis requires preservation of the modified RBC proteins, as post-translational modifications are non-durable. Moreover, classic protein-detection methods (e.g., western blotting) are notoriously difficult to standardize in RBCs due to the low abundance of proteins relative to hemoglobin, which accounts for ~98% of the protein content in these cells12. Thus, antibody-based staining of chemically-preserved RBCs has been the method of choice when investigating acute modifications of important RBC proteins, such as the RBC-specific isoform of nitric oxide synthase (RBC-NOS)13,14. RBC-NOS has been shown to enzymatically produce nitric oxide (NO), which seems indispensable for essential RBC properties, including RBC deformability15,16,17. Post-translational modifications of RBC-NOS regulate catalytic enzyme activity, with phosphorylation of the serine 1177 residue being described to increase enzyme activity, while phosphorylation of the residues serine 114 or threonine 495 have been linked with decreased RBC-NOS activity18,19.
Collectively, temporary modifications of RBC proteins contribute to important cellular function, and standardized protocols that enable detection of these modified proteins are of high value. Here, we present two distinct protocols that exploit specific antibodies to facilitate the detection of RBC-NOS protein activation, and discuss recommendations for data analysis and interpretation.
Performance of the described protocols was assessed by measuring the well-reported increase in the phosphorylation of RBC-NOS at the serine 1177 residue in response to mechanical forces reflective of those occurring within the human vasculature (5 Pa).
Protocol
The protocols described here are in alignment with the Declaration of Helsinki and were approved by the Ethics Committees of the German Sports University Cologne (9/16/2013) and Griffith University (2019/808). Volunteers were screened to ensure the absence of relevant pathologies and provided written informed consent.
1. Staining of RBC proteins using immunohistochemistry protocols
NOTE: A detailed list of the required chemicals and materials is provided in the Table of Materials. The following sections describe the preparation of the required solutions, followed by a detailed description of the immunohistochemistry protocol (Figure 1).
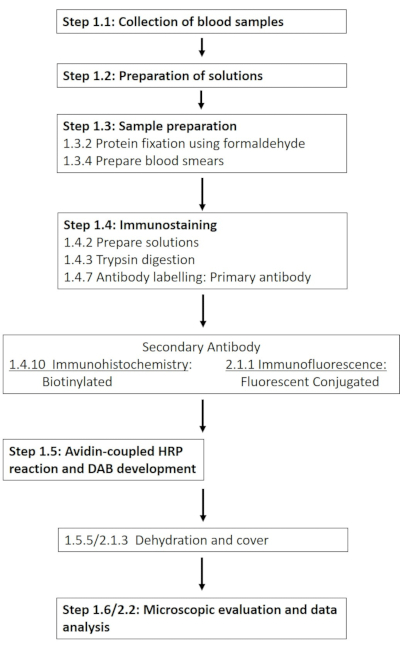
Figure 1: Schematic of the individual steps required for the immunohistochemical and immunofluorescence staining of RBC-NOS at phosphorylation site 1177. A typical workflow of the presented protocols stretching from solution preparation and blood sampling to antibody-based detection and visualization is presented. Please click here to view a larger version of this figure.
- Collection of blood sample
- Obtain blood samples (10 mL of whole blood) from seven healthy males. The volunteers were healthy individuals reportedly free from cardiovascular, hematological, neurological, endocrine, or metabolic diseases. The volunteers were also non-smokers.
- Collect blood using a sterile needle and syringe from a prominent vein in the antecubital region of the forearm, and immediately transfer into tubes coated with one of the following anticoagulants: sodium heparin (for immunohistochemistry) or ethylenediaminetetraacetic acid (EDTA; for fluorescent labeling).
- Expose the anticoagulated blood samples to precisely-controlled mechanical forces using a Couette-type shearing system, as described before7,20.
- The shearing apparatus comprises a rotatable cup and a stationary bob, separated by a 300 µm gap. Transfer the blood sample into the gap, which causes the precisely controllable rotational velocity of the cup to exert well-controlled mechanical forces onto the sample. Representative data presented here were produced by applying mechanical forces reflective of those occurring within the human vasculature (5 Pa) for extended durations (300 s).
- Preparation of solutions necessary for immunostaining
- Prepare the solutions as in Table 1. The solutions can be prepared prior to performing the immunostaining procedure and stored at given temperature until needed.
NOTE: Storage duration may vary between the chemicals. Check the material safety data sheet.
- Prepare the solutions as in Table 1. The solutions can be prepared prior to performing the immunostaining procedure and stored at given temperature until needed.
- Sample preparation
- Process whole blood immediately after withdrawal or experimental treatment (where applicable; e.g., mechanical stimulus in our samples; see step 1.1.3) to be able to detect short-living effects, for example after shear stress application, exercise, short-duration hypoxia exposure, etc.
- Perform RBC protein fixation using formaldehyde as follows: dilute whole blood at a ratio of 1:2 in 4% paraformaldehyde solution for 20 min at room temperature (RT). Centrifuge the sample at 132 x g for 3 min at RT and carefully remove the supernatant by pipetting.
- Re-suspend the RBC pellet in two volumes of 0.1 mol/L phosphate buffered saline (PBS) (1:3 dilution) and incubate for 5 min at RT. Repeat the centrifugation as described above and remove the supernatant, which should be clear, by pipetting. Re-suspend the RBC pellet in one volume of 0.1 mol/L PBS (1:2 dilution).
- Prepare blood smears as follows: label a microscope slide with the sample ID, antibody applied, etc. using an alcohol resistant pen (e.g., pencil). Then, add 10 µL of the prepared RBC solution right above the label field.
- Place a second slide on the sample at an angle of approximately 45° and disperse the sample evenly along the slide. Heat-fix the slide over a Bunsen burner by hovering the sample over the burner with constant movement for 5-7 s.
NOTE: It is recommended to air-dry the samples prior to heat fixing. The samples can be stored at RT until staining (Figure 2).
- Immunohistochemistry staining
- Mark two areas on each slide: a test area in which the RBCs are incubated with the respective primary and secondary antibody, and a control area in which the primary antibody is replaced by a control solution. This area serves as a within assay control to determine the background signal of the RBCs.
- Prepare solutions prior to or during the procedure, as per Table 2. A volume of 300 µL is needed per test area and 200 µL per control area.
NOTE: It is important to note that these antibody-based staining procedures yield semi-quantitative, rather than quantitative, data. Thus, to ensure that meaningful conclusions may be drawn from the produced data, appropriate control samples (i.e., to provide a reference point for the assessed relative changes in protein activation) are absolutely required. - For trypsin digestion, thaw 0.1% trypsin and equilibrate to RT. Mark two areas on each slide using a grease pencil: a test area (2/3 of the slide) and a control area (1/3 of the slide). Carefully apply tris-buffered saline (TBS) using transfer pipettes to wash the sample areas. Let the TBS sit for 30 s and then pour it off. Repeat the washing step.
NOTE: Add the following solutions to both the control and the test area, unless otherwise described. The areas need to be sufficiently covered with the respective solution. Apply the solutions using disposable transfer pipettes, and switch pipettes after each solution. - Add 0.1% trypsin, cover the slides using a purpose-built incubation chamber with either a lid or aluminum foil, and incubate for 30 min at 37 °C in an incubator. Following incubation, stop the enzyme reaction by adding tap water. Pour off the solution. Wash both areas 3x with TBS, as described above.
- For blocking peroxidase activation, add methanol solution and incubate for 30 min at RT. Cover the slides during that time. Pour off the solution.
NOTE: For immunofluorescence detection (section 2), this step may be omitted, given that the blocking of endogenous peroxidases serves to prevent artifactual signal caused by the detection with horseradish peroxidase (HRP) and 3,3′-Diaminobenzidine hydrate (DAB). - Wash both areas 3x with TBS, as described in step 1.4.3. Add 3% skim milk solution and incubate for 30 min at RT to block. Pour off the solution. Do not wash afterwards.
- Add primary antibody (AB) only to the test area. Add AB control solution (see Table 2) to the control area. Incubate samples at 4 °C overnight. Cover slides during that time to prevent drying.
NOTE: Avoid transferring the solutions from the test to the control area, as this will influence the control values. - Pour off the antibody solution. Wash both areas 3x with TBS, as described in step 1.4.3.
- Perform a blocking step to prevent unspecific binding. Add 3% normal goat serum and incubate for 30 min at RT. Pour off the solution.
- Add the secondary antibody solution and incubate for 30 min at RT. Pour off the antibody solution. Wash the areas 3x with TBS, as described in step 1.4.3.
- Development of staining and cover
- Carry out the avidin-coupled HRP reaction (see dilution in Table 2) by adding diluted avidin-peroxidase solution and incubating for 30 min at RT. Pour off the solution.
- Prepare the DAB mixture to develop the immunostaining prior to use, as per Table 3.
CAUTION: DAB is dangerous. Consider hazard statements H341 (suspected of causing genetic defects) and H350 (may cause cancer). Consider the following precautionary statements: P201, P202, P280, P308+P313, P405, and P501. - Perform DAB control staining as follows: collect the HRP solution from step 1.5.1 from a slide and mix with a small volume (e.g., 1 mL) of prepared DAB solution in a separate centrifuge tube before staining. The mixture should develop a brown/gray color.
- Place the slides under a microscope (magnification of at least 200x) and add DAB solution to both areas. Monitor the staining of the RBCs continuously and stop the staining by removing the DAB solution with a disposable pipette before the background starts to color. For RBC-NOS serine 1177 staining, the DAB incubation time is about 17 min.
- Perform sample dehydration. Place the slides in a glass rack and immerse for 5 s each in ethanol solutions of various dilutions, starting with 70%, followed by 96%, and then 100%, and finally in xylol. Remove the slides from the rack and place on a tissue, with RBCs at the top to absorb excess liquid.
- Add two or three drops of mounting medium throughout the slide. Cover the slide using a coverslip. Avoid the inclusion of air bubbles, as this hinders the microscopic evaluation. Dry the samples at least overnight in a fume hood.
- Performing microscopic evaluation
- For visualization and imaging, place slides in a transmitted-light microscope with a magnification of at least 200x. Ensure that the microscope is coupled to a camera to take pictures of the stained RBCs.
- Turn on the microscope light source. Turn on the microscope-attached camera and start the microscope control software. Use brightfield microscopy to determine the appropriate focus by turning the coarse and fine focus adjustment knobs and finding the level of focus where RBCs are visible.
- Set the background values of the pictures to 220 ± 5 gray values, measured on three cell-free areas of the slide. To do so, open the first picture using the freely available software ImageJ. Select the option Mean gray values using the panel Set measurements. Use the oval icon to measure the gray value using the Measure command. If the measured values are out of range, adjust the background light at the microscope accordingly. Repeat these steps until the background values are correct.
NOTE: These background values should be within the given range for all images taken. Otherwise, the data are not comparable. - For analysis of RBC gray value, mark the edge of each RBC using the Oval selection tool within the ImageJ software. Determine the gray values of individual RBCs using the Measure command. Measure the gray values of a minimum of 50 RBCs from the test and a minimum of 10 RBCs from the control area.
NOTE: The total number of analyzed RBCs can be adjusted individually. The more RBCs analyzed, the more meaningful the result. However, the total number of analyzed RBCs should be comparable for each condition, subject, etc. within a given experiment.- Avoid analysis of RBCs near the grease pencil, as staining might be incomplete in this area. Avoid analysis of overlapping RBCs, as this affects the staining intensity. Only use RBCs that are separated from other RBCs. Analyze at least five images from the test and at least two images from the control area for the evaluation of 50/10 RBCs. Include RBCs from every picture in the final analysis.
- To calculate staining intensity, compute the final signals as:
(individual test area RBC – mean test area background) – (individual control area RBC – mean control area background)
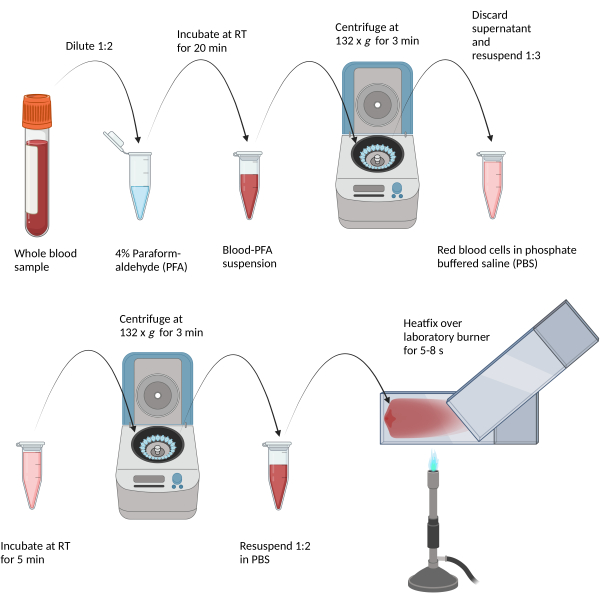
Figure 2: Depiction of the fixative process and blood smear generation. (Scheme created with BioRender.com.) Diluted blood samples are chemically fixed in paraformaldehyde, then centrifuged and washed with phosphate-buffered saline. Finally, resuspended blood is smeared onto a glass slide and thermally fixed via hovering over a Bunsen burner flame. Please click here to view a larger version of this figure.
Table 1: Preparation and storage conditions for solutions necessary for immunohistochemical staining. The solution can be prepared prior to the protocol. Please click here to download this Table.
Table 2: Description of antibody solutions for immediate use. Please click here to download this Table.
Table 3: Components and protocol to prepare DAB solution for immediate use. Please click here to download this Table.
2. Fluorescent labelling of RBC proteins
NOTE: The following section outlines an adaptation of the immunohistochemical protocol, developed with the aim of enabling the use of antibodies with fluorescent conjugates (Figure 1).
Blood sample preparation for the immunofluorescence protocol is identical to that described in section 1, so the following section commences from the staining of samples.
- Immunofluorescence staining
- Incubate with the secondary, fluorescent conjugated antibody (for the volume, see step 1.4.2) for 30 min at RT protected from light, using either a purpose-built incubation chamber or aluminum foil.
- Pour off the secondary antibody solution and wash the samples 3x with TBS. Leave the final wash on the samples to prevent them from drying out.
- Pour off the TBS and remove the samples from the sample rack one at a time to prevent prolonged light exposure. For dehydrating the samples, expose the slides to different ethanol solutions for ~5 s each, starting with 70%, followed by 90%, and finally 100%. Expose the slides to xylol/xylene solution for 5 s.
- Prepare a coverslip with two or three drops of mounting medium and then mount the slide with the coverslip. Ensure even distribution of the mounting medium by applying light pressure with tweezers or a similar, sterile metal instrument. Eliminate air bubbles, which may otherwise interfere with imaging, with the same instrument. Leave the samples to dry overnight in a dark and dry space.
- Microscopic evaluation
- For visualization and imaging, place the slide on the microscope stage with a total magnification of at least 400x. Turn on the microscope light source and the fluorescent light source, ensuring that the fluorescent light source is adjusted to maximal intensity.
- Turn on the microscope-attached camera and start the microscope control software. Use brightfield microscopy to determine the appropriate focus by turning the coarse and fine focus adjustment knobs, and finding the level of focus where RBCs are visible.
NOTE: Excessive exposure to light may cause photobleaching of the fluorescent conjugates. One may use a slide from a previous experiment for this purpose, to minimize light exposure of the slides that are yet to be analyzed. - Determine optimal laser intensity by inspecting RBCs in the control area. Ensure that the intensity is high enough that RBCs are visible, while not producing a large background signal. Keep the intensity and exposure time consistent between areas and samples to ensure comparability.
- Capture brightfield and fluorescent images in at least three distinct areas of the test area of the slide, selected at random. To select an area, pan away from the edges of the slide that are marked by the lipid pen, using the microscope stage controls. Select an area that exhibits uniform distribution of a singular RBC layer.
- Set the exposure time to 1 s for fluorescent images and capture an image using the software controls of the microscope control software. Switch to brightfield mode, set the exposure time to 'Auto', and capture the corresponding brightfield image.
- Capture brightfield and fluorescent images in at least two distinct randomly selected areas of the control area of the slide by repeating step 2.2.3.
- Save the images in .tif format to preserve the original gray values of pixels, prevent compression, and carry metadata of the acquisition.
- Measure the gray values of the captured RBCs. Open ImageJ (or FIJI21; see Supplementary File 1). Determine the gray values of individual RBCs. To this end, mark each individual cell with the Oval selection tool, and analyze using the Measure command.
NOTE: RBCs should not be analyzed if they overlap with other cells, as this may increase the resultant signal. - Highlight between three and five areas free of RBCs and determine the gray values to provide a measure of background signal. Analyze at least 150 RBCs from at least three distinct images for each test area, and at least 50 RBCs from at least two distinct images for each control area.
NOTE: Analysis of more cells/areas may be advised to minimize variability. Given that the RBC population is inherently heterogenous, cell-cell variability in signal may be significant, depending on the protein of interest. - Perform alternative data analysis of captured images using macro command. Create/install a macro command through the FIJI release of ImageJ for automated selection, background correction, and gray value analysis of a given image.
NOTE: This routine uses automated thresholding to detect the cells present in a given image using a copy of the original image, producing an overlay that is imposed onto the original image to extract gray values of fluorescent RBCs. The macro is deposited as a .ijm file, as Supplementary Coding File 1. - Open the image file in .tif format ready for analysis in FIJI. Open the RBC fluorescence.ijm (Supplementary Coding File 1) macro and click Run.
NOTE: The macro is set up for images obtained with 600x magnification and a large signal-to-noise ratio. Automatic selection of cells should be reviewed by the investigator.
- Compute the final signals as:
(test area RBC – test area background) – (control area RBC – control area background) for manual analysis;
(test area – control area) for automated analysis.
Representative Results
The presented protocol, describing methods that facilitate the detection of acute alterations in RBC proteins, was tested on a well-known mechanically sensitive protein alteration: phosphorylation of RBC-NOS at the serine 1177 residue. Whole blood was obtained from healthy volunteers and subsequently split into two separate aliquots. A given blood sample was exposed to mechanical shear stress of physiological magnitude (5 Pa) for 300 s, which was previously shown to elicit RBC-NOS phosphorylation at serine 117714. Immediately following cessation of the mechanical shear exposure, the blood sample was fixed in paraformaldehyde. As the control, a blood sample was exposed, loaded into the shearing device, and left to rest for 300 s prior to fixation in paraformaldehyde. The signal of antibodies targeted against RBC-NOS phosphorylated at the serine 1177 residue at rest and in response to mechanical force exposure was assessed using both immunohistochemistry (Figure 3A,B) and immunofluorescence (Figure 3C,D). Sheared and non-sheared samples produced statistically significantly different signals [Friedman test: χ2(3) = 18.71, p = 0.0003]. A comparable, approximately threefold increase in the signal of antibodies targeted against RBC-NOS phosphorylated at the serine 1177 residue was detected in response to mechanical force exposure of RBCs, compared to the respective unsheared cells (both p < 0.01) when assessed with either the immunohistochemical or immunofluorescence protocol (Figure 3E).
Thus, comparison of the results obtained with both methods indicates excellent agreement between the presented protocols, which also successfully and reliably detected increased RBC-NOS phosphorylation in response to mechanical stimulation.
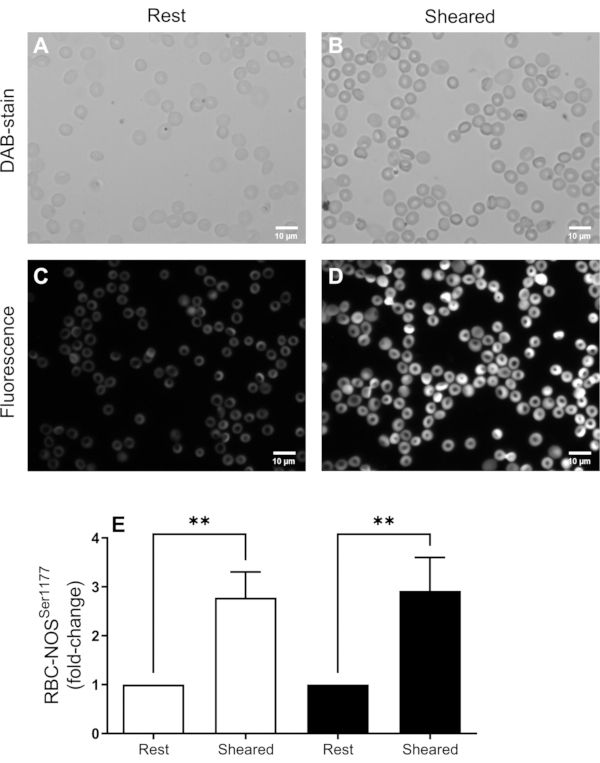
Figure 3: Representative pooled data from immunohistochemical (A,B) and immunofluorescent (C,D) staining of RBC-NOS serine 1177 phosphorylation following mechanical shear. Analysis of the signal intensity from these samples is presented in (E), where white bars reflect data obtained using HRP staining, and black bars represent data obtained using the fluorescent method. Blood was either prepared at rest or immediately following exposure to mechanical force (i.e., shear). N = 7 blood samples were obtained from distinct donors. Data shown as mean ± standard error of the mean. **p < 0.01, determined using a non-parametric Friedman test. Please click here to view a larger version of this figure.
Supplementary File 1: Automated semi-quantitative image analysis raw code with step-by-step annotations for images of immunofluorescent red blood cells. Please click here to download this File.
Supplementary Coding File 1: Compiled code compatible with FIJI/ImageJ-software to run automated image analysis of immunofluorescent red blood cells. Please click here to download this File.
Discussion
Recent literature highly suggests that the RBC-NOS protein is of crucial importance for the regulation of RBC deformability15,22,23, which in turn facilitates their passage through narrow capillaries24. Protein activity highly depends on post-translational protein modifications, particularly the phosphorylation of certain residues18. The focus of interest lies in the phosphorylation site 1177, which relates to the activation of the RBC-NOS protein23. Alterations of this protein have been shown in a variety of diseases25,26,27,28,29, thus, investigation of these changes could provide valuable knowledge not only for the understanding of certain diseases, but also to develop and guide specific therapies30.
Several stimuli have been identified to increase RBC-NOS serine 1177 phosphorylation22, but mechanical forces appear to be a major extracellular stimulus that causes activation or modulation of the RBC-NOS protein13,18,31,32. However, modulations of RBC-NOS activity are rather temporary33; thus, analysis/preservation of shear-dependent alterations must be performed immediately. Immunohistochemical, and later immunofluorescence protocols, were developed to facilitate preservation and analysis of acute, regulatory modifications of RBC-NOS.
The results presented here, obtained via immunohistochemical and immunofluorescence protocols, show a high level of agreement regarding the increased phosphorylation of RBC-NOS serine 1177 after shear exposure. Thus, both protocols are suitable for the investigation of transient post-translational changes of RBC proteins, and the respective experimenter can decide which method they might use in their respective laboratory, considering the locally available resources and infrastructure. Given the wide commercial availability of antibodies targeting proteins, both in their native state or following post-translational modifications, the present assays are adaptable for a considerable range of targets. Thus, they present useful tools for assessing RBC signaling14,27,34.
Certain aspects should be considered during the procedure. The mentioned antibodies are not specifically developed to be applied in RBCs. Instead, these are specific endothelial-type NOS (eNOS) antibodies. Since eNOS and RBC-NOS seem to share great homology22,23, eNOS-specific antibodies have traditionally been used to visualize RBC-NOS activation. It is important to note that antibodies directed against the inducible (iNOS) or neuronal (nNOS) isoforms do not produce signals in RBCs, supporting that RBC-NOS shares significant structural resemblance with eNOS22. The appropriate dilution must be tested before any antibody is used in the experiment, since the supplier-recommended data cannot be applied to RBC experimentation without testing. Further, a switch of distributing companies requires excessive testing of various dilutions prior to usage. We recommend following a highly systemic approach when testing newly sourced antibodies/chemicals; new components should be tested with dose-response approaches, where only the specific steps introducing the new component should be altered. Positive controls (i.e., stimulating RBC-NOS activation) should be provided by comparing sheared with unsheared blood, as presented here. Alternatively, pharmacological stimulation of RBC-NOS phosphorylation with 350 pM insulin has been shown to result in increased RBC-NOS phosphorylation, similar to that observed with mechanical force application7.
The limitations of the presented detection methods extend to common limitations of methods that rely on the specificity of commercially obtained primary antibodies (e.g., western blot). First, a primary antibody targeted to the antigen of interest must be available. If available, the antibody must be specific for the target, which may be confirmed by functional measures (i.e., pharmacological treatment with activators/inhibitors) or using western blotting. Moreover, the data produced via the described methods must be carefully interpreted. That is, the presented methods are semi-quantitative, providing information on changes in protein modifications relative to appropriate control samples. The semi-quantitative evaluation presented here always refers to the activation of the enzyme, and should not be confused with the enzyme activity. Measurements of enzyme activity require separate assays, such as the arginine-citrulline assay which measures the rate of conversion (fmol/min) of [3H] L-arginine to [3H] citrulline or [14C] L-arginine to [14C] citrulline35,36. To verify whether an increased RBC-NOS activation is also accompanied by higher NO levels and/or increased RBC deformability values, additional analysis of the NO concentration, for example, should be performed. Moreover, to single out RBC-NOS as a source of NO, NOS inhibitors such as L-NIO should be employed15.
It can be summarized that the methods presented here are well-developed to analyze changes in RBC-NOS activation in response to applied stimuli relative to unstimulated control cells. These methods thus contribute to the understanding how mechanical stress affects the functions of RBC proteins, which are critical to cellular function, and ultimately vital to adequate perfusion of working tissue and gas exchange.
開示
The authors have nothing to disclose.
Acknowledgements
LK acknowledges the support of an Australian Government Research Training Program Scholarship.
Materials
| 3,3′-Diaminobenzidin -tetrahydrochloride Hydrate | Sigma/Merck | D5637 | DAB |
| Ammoniumchloride | Merck /Millipore | 101145 | NH4Cl |
| Centrifuge 5427 R | Eppendorf | 5409000010 | |
| Coverslips | VWR | 631-0147 | |
| di-sodium Hydrogen Phosphate Dihydrate | Merck /Millipore | 106580 | Na2HPO4. 2 H2O |
| Disposable transfer pipettes | VWR | 612-6803 | |
| Entellan | Merck /Millipore | 107961 | rapid mounting medium for microscopy |
| Ethanol denaturated using 1 % methyl ethyl ketone (MEK) | Hofmann | 642 | |
| Glucose-Oxidase | Sigma/Merck | G2133 | |
| Grease pencil | Dako | S 2002 | |
| Horse-radish peroxidase/ExtrAvidin−Peroxidase | Sigma/Merck | E-2886 | HRP |
| Hydrochloric acid | Merck /Millipore | 109057 | HCl |
| Hydrogen peroxide, 30% | Merck /Millipore | 107203 | H2O2 |
| ImageJ Software | Freeware | ||
| Laser-assisted optical rotational cell analyser (LORCA) | RR Mechatronics | Ektacytometer instrument used for shearing | |
| Methanol | Merck /Millipore | 106009 | |
| Microscope slides | VWR | 630-1985 | |
| Nickel(II)-sulfate Hexahydrate | Sigma/Merck | N4882 | NiSO4.6H2O |
| Normal Goat serum | Agilent/DAKO | X0907 | NGS |
| Paraformaldehyde | Merck /Millipore | 818715 | PFA |
| Pipettes Eppendorf Reference 2 | VWR | 613-5836/ 613-5839 | |
| Rabbit Anti-phospho eNOS Antibody (Ser1177) | Merck/Millipore | 07-428-I | Primary Antibody |
| Reaction tubes, 2ml | Eppendorf | 30120094 | |
| Secondary Antibody goat anti rabbit | Agilent/DAKO | E0432 | Secondary Antibody |
| Skim milk powder | Bio-Rad | 170-6404 | |
| Sodium chloride | Merck /Millipore | 106404 | NaCl |
| Sodium Dihydrogen Phosphate Monohydrate | Merck /Millipore | 106346 | NaH2PO4.H2O |
| Sodium hydroxide, 1 M | Merck /Millipore | 150706 | NaOH |
| Tris(hydroxymethyl)-aminomethane | Merck /Millipore | 108382 | Tris |
| Trypsin | Sigma/Merck | T7409 | |
| Tween20 | Merck /Millipore | 822184 | |
| Whatman Glas microfiber filter, quality GF/F | Merck /Millipore | WHA1825047 | |
| Xylol | VWR Chemicals | 2,89,73,465 | |
| ß-D-Glucose monohydrate | Merck /Millipore | 14431-43-7 |
参考文献
- Cohen, R. M., et al. Red cell life span heterogeneity in hematologically normal people is sufficient to alter HbA1c. Blood. 112 (10), 4284-4291 (2008).
- Mock, D. M., et al. Red blood cell (RBC) survival determined in humans using RBCs labeled at multiple biotin densities. Transfusion. 51 (5), 1047-1057 (2011).
- Thiagarajan, P., Parker, C. J., Prchal, J. T. How do red blood cells die?. Frontiers in Physiology. 12, 655393 (2021).
- Moras, M., Lefevre, S. D., Ostuni, M. A. From erythroblasts to mature red blood cells: organelle clearance in mammals. Frontiers in Physiology. 8, 1076 (2017).
- Pretini, V., et al. Red blood cells: chasing interactions. Frontiers in Physiology. 10, 945 (2019).
- Cahalan, S. M., et al. Piezo1 links mechanical forces to red blood cell volume. eLife. 4, e07370 (2015).
- Kuck, L., Peart, J. N., Simmonds, M. J. Piezo1 regulates shear-dependent nitric oxide production in human erythrocytes. American Journal of Physiology. Heart and Circulatory Physiology. 323 (1), H24-H37 (2022).
- Kuck, L., Peart, J. N., Simmonds, M. J. Active modulation of human erythrocyte mechanics. American Journal of Physiology. Cell Physiology. 319 (2), C250-C257 (2020).
- Strader, M. B., et al. Post-translational modification as a response to cellular stress induced by hemoglobin oxidation in sickle cell disease. Scientific Reports. 10 (1), 14218 (2020).
- Pecankova, K., Majek, P., Cermak, J., Dyr, J. E. Posttranslational modifications of red blood cell ghost proteins as "signatures" for distinguishing between low- and high-risk myelodysplastic syndrome patients. Turkish Journal of Haematology. 34 (1), 111-113 (2017).
- Grau, M., et al. High red blood cell nitric oxide synthase activation is not associated with improved vascular function and red blood cell deformability in sickle cell anaemia. British Journal of Haematology. 168 (5), 728-736 (2015).
- Sae-Lee, W., et al. The protein organization of a red blood cell. Cell Reports. 40 (3), 111103 (2022).
- Suhr, F., et al. Moderate exercise promotes human RBC-NOS activity, NO production and deformability through Akt kinase pathway. PLoS One. 7 (9), e45982 (2012).
- Kuck, L., Grau, M., Bloch, W., Simmonds, M. J. Shear stress ameliorates superoxide impairment to erythrocyte deformability with concurrent nitric oxide synthase activation. Frontiers in Physiology. 10, 36 (2019).
- Grau, M., et al. RBC-NOS-dependent S-nitrosylation of cytoskeletal proteins improves RBC deformability. PLoS One. 8 (2), e56759 (2013).
- Simmonds, M. J., Detterich, J. A., Connes, P. Nitric oxide, vasodilation and the red blood cell. Biorheology. 51 (2-3), 121-134 (2014).
- Bor-Kucukatay, M., Wenby, R. B., Meiselman, H. J., Baskurt, O. K. Effects of nitric oxide on red blood cell deformability. American Journal of Physiology. Heart and Circulatory Physiology. 284 (5), H1577-H1584 (2003).
- Suhr, F., et al. Intensive exercise induces changes of endothelial nitric oxide synthase pattern in human erythrocytes. Nitric Oxide: Biology and Chemistry. 20 (2), 95-103 (2009).
- Grau, M., et al. Regulation of red blood cell deformability is independent of red blood cell-nitric oxide synthase under hypoxia. Clinical Hemorheology and Microcirculation. 63 (3), 199-215 (2016).
- Grau, M., Kuck, L., Dietz, T., Bloch, W., Simmonds, M. J. Sub-fractions of red blood cells respond differently to shear exposure following superoxide treatment. 生物学. 10 (1), 47 (2021).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Ozüyaman, B., Grau, M., Kelm, M., Merx, M. W., Kleinbongard, P. RBC NOS: regulatory mechanisms and therapeutic aspects. Trends in Molecular Medicine. 14 (7), 314-322 (2008).
- Kleinbongard, P., et al. Red blood cells express a functional endothelial nitric oxide synthase. Blood. 107 (7), 2943-2951 (2006).
- McMahon, T. J. Red blood cell deformability, vasoactive mediators, and adhesion. Frontiers in Physiology. 10, 1417 (2019).
- Bizjak, D. A., Brinkmann, C., Bloch, W., Grau, M. Increase in red blood cell-nitric oxide synthase dependent nitric oxide production during red blood cell aging in health and disease: a study on age dependent changes of rheologic and enzymatic properties in red blood cells. PLoS One. 10 (4), 0125206 (2015).
- Di Pietro, N., et al. Nitric oxide synthetic pathway and cGMP levels are altered in red blood cells from end-stage renal disease patients. Molecular and Cellular Biochemistry. 417 (1-2), 155-167 (2016).
- Grau, M., et al. Even patients with mild COVID-19 symptoms after SARS-CoV-2 infection show prolonged altered red blood cell morphology and rheological parameters. Journal of Cellular and Molecular Medicine. 26 (10), 3022-3030 (2022).
- Mozar, A., et al. Red blood cell nitric oxide synthase modulates red blood cell deformability in sickle cell anemia. Clinical Hemorheology and Microcirculation. 64 (1), 47-53 (2016).
- Ulker, P., Gunduz, F., Meiselman, H. J., Baskurt, O. K. Nitric oxide generated by red blood cells following exposure to shear stress dilates isolated small mesenteric arteries under hypoxic conditions. Clinical Hemorheology and Microcirculation. 54 (4), 357-369 (2013).
- Nader, E., et al. Hydroxyurea therapy modulates sickle cell anemia red blood cell physiology: Impact on RBC deformability, oxidative stress, nitrite levels and nitric oxide synthase signalling pathway. Nitric Oxide: Biology and Chemistry. 81, 28-35 (2018).
- Fischer, U. M., Schindler, R., Brixius, K., Mehlhorn, U., Bloch, W. Extracorporeal circulation activates endothelial nitric oxide synthase in erythrocytes. The Annals of Thoracic Surgery. 84 (6), 2000-2003 (2007).
- Horobin, J. T., Sabapathy, S., Kuck, L., Simmonds, M. J. Shear stress and RBC-NOS Serine1177 Phosphorylation in humans: a dose response. Life. 11 (1), 36 (2021).
- Kuck, L., Grau, M., Simmonds, M. J. Recovery time course of erythrocyte deformability following exposure to shear is dependent upon conditioning shear stress. Biorheology. 54 (5-6), 141-152 (2018).
- Grau, M., et al. Effect of acute exercise on RBC deformability and RBC nitric oxide synthase signalling pathway in young sickle cell anaemia patients. Scientific Reports. 9 (1), 11813 (2019).
- Feelisch, M. . Methods in Nitric Oxide Research. , (1998).
- Cortese-Krott, M. M., et al. Human red blood cells at work: identification and visualization of erythrocytic eNOS activity in health and disease. Blood>. 120 (20), 4229-4237 (2012).

