A Strategy to Identify de Novo Mutations in Common Disorders such as Autism and Schizophrenia
Summary
Molecular genetic strategy for finding de novo mutations causing common disorders such as autism and schizophrenia.
Abstract
There are several lines of evidence supporting the role of de novo mutations as a mechanism for common disorders, such as autism and schizophrenia. First, the de novo mutation rate in humans is relatively high, so new mutations are generated at a high frequency in the population. However, de novo mutations have not been reported in most common diseases. Mutations in genes leading to severe diseases where there is a strong negative selection against the phenotype, such as lethality in embryonic stages or reduced reproductive fitness, will not be transmitted to multiple family members, and therefore will not be detected by linkage gene mapping or association studies. The observation of very high concordance in monozygotic twins and very low concordance in dizygotic twins also strongly supports the hypothesis that a significant fraction of cases may result from new mutations. Such is the case for diseases such as autism and schizophrenia. Second, despite reduced reproductive fitness1 and extremely variable environmental factors, the incidence of some diseases is maintained worldwide at a relatively high and constant rate. This is the case for autism and schizophrenia, with an incidence of approximately 1% worldwide. Mutational load can be thought of as a balance between selection for or against a deleterious mutation and its production by de novo mutation. Lower rates of reproduction constitute a negative selection factor that should reduce the number of mutant alleles in the population, ultimately leading to decreased disease prevalence. These selective pressures tend to be of different intensity in different environments. Nonetheless, these severe mental disorders have been maintained at a constant relatively high prevalence in the worldwide population across a wide range of cultures and countries despite a strong negative selection against them2. This is not what one would predict in diseases with reduced reproductive fitness, unless there was a high new mutation rate. Finally, the effects of paternal age: there is a significantly increased risk of the disease with increasing paternal age, which could result from the age related increase in paternal de novo mutations. This is the case for autism and schizophrenia3. The male-to-female ratio of mutation rate is estimated at about 4–6:1, presumably due to a higher number of germ-cell divisions with age in males. Therefore, one would predict that de novo mutations would more frequently come from males, particularly older males4. A high rate of new mutations may in part explain why genetic studies have so far failed to identify many genes predisposing to complexes diseases genes, such as autism and schizophrenia, and why diseases have been identified for a mere 3% of genes in the human genome. Identification for de novo mutations as a cause of a disease requires a targeted molecular approach, which includes studying parents and affected subjects. The process for determining if the genetic basis of a disease may result in part from de novo mutations and the molecular approach to establish this link will be illustrated, using autism and schizophrenia as examples.
Protocol
1. Selection of disease that may be caused by de novo mutations
A disease that corresponds to the following criteria can fit with the de novo mutation hypothesis:
- The reproductive fitness is reduced.
- The frequency of the disease is relatively high and constant despite widely varying environments.
- The disease is associated with a higher paternal age.
- The classic linkage and association studies failed to explain a significant fraction of the disease heritability.
- The twin concordance data support a de novo model.
Analysis of the likelihood that a common disease where de novo mutations may in part explain the genetic basis is a critical first step.
2. Selection of cases and DNA samples
Selection of appropriate samples is critical for the success of the identification of de novo mutations. To maximize the chance of finding de novo mutations, we recommend the following:
- Select cases with early age of onset, severe phenotype, with unaffected parents, older fathers and with no extended family history of the disease.
- Choose patients whose available DNAs are sufficient to conduct the study. Especially critical is the availability of DNA from a primary cell source that was not subjected to culturing (e.g. Blood DNA or saliva DNA),
- The availability of both parents DNA is critical in order to determine the mutation transmission status (inherited vs. de novo). Availability of additional affected cohorts and normal controls is necessary for genetic validation studies once candidate genes are identified.
- Estimate the sample size based on the mutations rate, the amount of genes to be screened and the estimate of the fraction of cases that may result from a de novo mutation.
3. Gene resequencing; two major approaches
- High quality low throughput sequencing
This approach is based on the candidate genes approaches.- Selection of candidate gene(s)
Select the best candidate genes based on a scoring system which is built on 6 major criteria. Then calculate the total that corresponded to the sum of all the points attributed using the six criteria listed in Table 1. See example from our project in figure 1 of selected and not selected genes distribution.

Table 1. Criteria used for the candidate gene selection
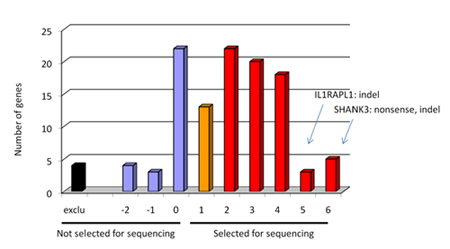
Figure 1. Graph showing the distribution of selected and not selected genes for sequencing in our project. We obtained a distribution of genes ranked by candidate properties by sorting genes according to their score value. For example, SHANK3 and NRXN1 genes, two genes that we found de novo mutation, had a score 7 and 6 respectively (maximum is 12). - Design primers using Primers3 software through Exonprimer. Only coding region and splice junction should be covered including an extra 50 base pairs on each side of the exon.
- Optimize PCR conditions for the choice of Taq, reaction volume, etc.
- Optimize all PCR fragments
- Amplify 5 ng of genomic DNA extracted from blood samples according to standard procedures
- Before sending for sequencing, do quality control of your PCR products by loading a 2% agarose gel. Selected randomly samples.
- Sequence the PCR products on a DNA Analyzer on one strand. A fragment is considered successfully sequenced if the analysis of over 90% of the traces is possible. This is applicable for a large scale screening.
- Variants Detection
- Use tools for detection and genotyping of genomic variations such as PolyPhred, Polyscan and Mutation Surveyor. A combination of more than 2 detection tools is ideal. For example, PolyPhred v.5 and PolyPhred v.6 with the default settings do not detect the same variations. Polyscan v.3 has a higher false positive mutation rate for SNPs (96%) and less for the INDELs (93%). PolyPhred v.6 did not detected the majority of true INDELs but have a false positive mutation rate (for INDELs) lower than Polyscan v.3 overall (90%). We should remove the option of SNPs detection for Polyscan v.3 and keep both PolyPhred v.5 and PolyPhred v.6 for variant detection. Mutation Surveyor and Polyscan are better for detecting indels. Note: The option of SNP detection should not be applied when using Polyscan. Only the “indel” option should be on. Polyscan generated to many false positive for SNP detection.
- For each unique novel exonic variants detected, confirm it manually by reamplifying the fragment and resequencing the proband and both parents using reverse and forward primers to eliminate any technical artifact.
- Selection of candidate gene(s)
- Whole exome sequencing
This approach is a high throughput sequencing targeting the majority of coding regions of the human genomes. We are now currently using this new approach in our lab, accelerating the detection of potential candidate genes.- Order the “SureSelect Human All Exon” targeting the coding region of over 16,000 genes (50 MB of the genome) designed by Agilent or any other similar product by others like Roche . Prior to order the capture kit, the sequencing platforms should be determine (Illumina vs SOLiD).
- Do the capture according to the Agilent protocol
- Sequence the product on your respective available next-generation platform
- Variant detection from the whole exome sequencing: Several bioinformatics tools for detection and genotyping of genomic variations from the next-generation sequencing platform are available such as BWA, Bfast, Bioscope which will perform the alignment. After which additional freely available downstream tools (for example SAM tools, Varscan, Annovar) would be needed it to call and annotate the variants. Commercial software that incorporates sequence alignment and variant calling and annotation are also available such as, NextGEN (Softgenetics), CLC Bio, and others
4. Genomic variants prioritization
Identified variants are then prioritized for follow up according to their probability in being de novo and deleterious to protein or mRNA function and /or structure. The variant follow up priorities for detection of de novo variant should be as follow:
- Unique variations (observed once in a single case)
- Variations not present in the parents
- Protein-truncating variations: nonsense, indels leading to frameshift and splicing mutations.
- Missense and silent variation predicted to be functionally disruptive (e.g., affect mRNA splicing). Use Polyphen, SIFT and PANTHER for functional prediction effect on the protein.
If using whole exome sequencing, selection of candidate genes can be used as a strategy for prioritizing variants for further study.
5. Genetic validation
- Resequence the entire gene in additional patient cases (to identify other causative mutations) and in controls. The control samples will be used to evaluate allelic frequencies of prioritized variants. Any gene containing at least two different de novo deleterious mutations (nonsense, splicing, frameshifts, and predicted damaging missense) found in different patients (but not in control samples or public databases) should be highly prioritized for further validation studies. This includes: 1) testing for a potential splicing defect in lymphoblastoid cell lines derived from the patient. In our experience, most genes yield RT-PCR products from lymphoblastoid cell lines; 2) investigate altered protein expression levels by quantitative Western blot analysis of protein extracts from the lymphoblastoid cell lines and 3) further test the mutation/gene at the functional level in animal (c elegans, Zebrafish) and cell line models as previously done by our group (ex: 5,6).
6. Representative Results:
Following this protocol, we were able to identify new genes for schizophrenia and autism. One example is our recently SHANK3 gene discovery (Figure 2). Two different de novo mutations in SHANK3 gene, one nonsense mutation found in three affected brother and one missense mutation in one affected female.
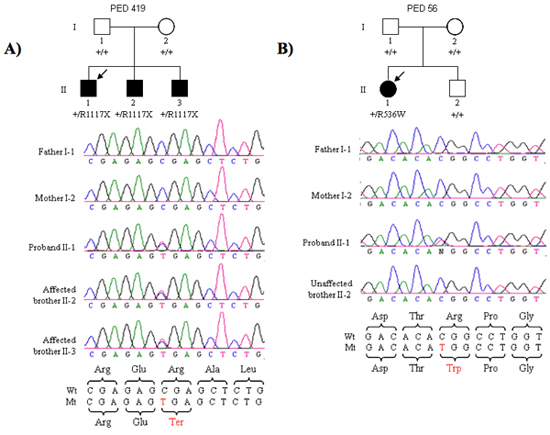
Figure 2. (A) Segregation of the R1117X nonsense mutation in three affected brothers of family PED 419. The proband is indicated by the arrow. (B) Segregation of the R536W missense mutation in the proband but not her non-affected brother in PED 56.
Discussion
The procedure outlined here aims to identify specific common diseases that likely result, in part, from de novo mutations, and to prove this hypothesis. De novo mutations are a well established mechanism for the development of a number of diseases, for example the hereditary cancer syndromes, but has been poorly explored in common diseases. This in part results from the technical challenges involved in the identification of de novo mutations, which requires the sequencing of large amounts of DNA, which has only very recently become cost effective with the advent of Next Generation Sequencing. In addition, the de novo mutation rate in humans was, until very recently, only an estimate. Only very recently have there been reports directly determining the mutation rate in humans. Prior to these measurements, it was difficult to predict the sample size needed for this kind of study and to determine if the observed de novo mutation rate is greater than the baseline rate. Sequencing candidate genes versus whole genome? Since the majority of reported disease mutations are missense/nonsense mutations and are splice site mutations (according to HGMD web site) our screening strategy would identify over 68% of known mutations. There is also a clear relationship between the severity of amino acid replacement and the likelihood of a clinical phenotype. As compared with a conservative amino acid substitution, a nonsense change is 9.0 times more likely to present clinically 7. Thus, at this time sequencing candidate genes is the most cost effective strategy.
The success of the outlined procedure depends on several critical steps, which are outlined in detail and illustrated using two examples, autism and schizophrenia. There are many pitfalls which need to be avoided, such as which disease to select, which patients to screen, source of DNA, and details of how to efficiently identify the de novo mutations. We provide a method for most efficiently determining the fraction of cases of any disease which results from such spontaneous mutations.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thanks our funding sources Genome Canada and Génome Québec, and Université de Montréal as well as funding from the Canadian Foundation for Innovation for funding our ‘Synapse to Disease’ (S2D) project.
Riferimenti
- Bassett, A. S., Bury, A., Hodgkinson, K. A., Honer, W. G. Reproductive fitness in familial schizophrenia. Schizophr Res. 21, 151-160 (1996).
- Jablensky, A. Schizophrenia: manifestations, incidence and course in different cultures. A World Health Organization ten-country study. Psychol Med Monogr. , 1-97 (1992).
- Malaspina, D. Schizophrenia risk and paternal age: a potential role for de novo mutations in schizophrenia vulnerability genes. CNS Spectr. 7, 26-29 (2002).
- DeLisi, L. E. A genome-wide scan for linkage to chromosomal regions in 382 sibling pairs with schizophrenia or schizoaffective disorder. Am J Psychiatry. 159, 803-812 (2002).
- Gauthier, J. De Novo SHANK3 Mutations in Patients Ascertained for Schizophrenia. Proc Natl Acad Sci U S A. , (2010).
- Piton, A. Mutations in the calcium-related gene IL1RAPL1 are associated with autism. Hum Mol Genet. 17, 3965-3974 (2008).
- Krawczak, M., Ball, E. V., Cooper, D. N. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet. 63, 474-488 (1998).

