Drosophila Embryo Preparation and Microinjection for Live Cell Microscopy Performed using an Automated High Content Analyzer
Summary
Presented here is a protocol to microinject and simultaneously image multiple Drosophila embryos during embryonic development using a plate-based, high content imager.
Abstract
Modern approaches in quantitative live cell imaging have become an essential tool for exploring cell biology, by enabling the use of statistics and computational modeling to classify and compare biological processes. Although cell culture model systems are great for high content imaging, high throughput studies of cell morphology suggest that ex vivo cultures are limited in recapitulating the morphological complexity found in cells within living organisms. As such, there is a need for a scalable high throughput model system to image living cells within an intact organism. Described here is a protocol for using a high content image analyzer to simultaneously acquire multiple time-lapse videos of embryonic Drosophila melanogaster development during the syncytial blastoderm stage. The syncytial blastoderm has traditionally served as a great in vivo model for imaging biological events; however, obtaining a significant number of experimental replicates for quantitative and high-throughput approaches has been labor intensive and limited by the imaging of a single embryo per experimental repeat. Presented here is a method to adapt imaging and microinjection approaches to suit a high content imaging system, or any inverted microscope capable of automated multipoint acquisition. This approach enables the simultaneous acquisition of 6-12 embryos, depending on desired acquisition factors, within a single imaging session.
Introduction
Over the past 30 years, advances in imaging technologies and molecular probes for live cell imaging have advanced our understanding of the complexity and inner workings of the cell. Accompanying this headway in technology is a greater reliance on the use of ex vivo cell culture models for the acquisition of high throughput imaging data. Cell culture models provide several advantages, including control of the microenvironment through microfluidic systems, and the ability to image multiple cells simultaneously, enabling the use of quantitative approaches necessary for high-throughput screening1,2. However, there are also significant disadvantages associated with cell culture models in the context of morphological studies, such as two-dimensional glass or plastic surfaces producing confounding effects on cell morphology and its regulation3,4,5. These effects can provide false or misleading data with regards to the study of biological processes regulating cellular morphology.
Although the alternative has been to study cellular morphology in the context of an intact organism6, few suitable intact organisms facilitate high throughput quantitative live cell imaging due to difficulties in keeping whole organisms alive through out imaging, and challenges associated with imaging inside a large multicellular organism. As a result, cell biological studies in intact organisms have generally focused on observing a single organism over time, which greatly limits sample size. In addition, this one-animal-at-a-time limitation makes live imaging difficult and costly to employ in the form of a screen and restricts the ability to apply quantitative analytical tools since the number of samples is generally too small. Thus, arises the need for a multicellular model organism that can be used for high content based quantitative approaches.
This protocol adapts the Drosophila embryo for plate-based high content imaging to generate experimental sample sizes large enough for quantitative analysis. Drosophila melanogaster is commonly known as the first model organism. Research using Drosophila has led to breakthroughs in cell and molecular biology, including the transmission of genetic information, identification of cell signaling pathways, and genetic requirements of embryonic development7,8,9. In addition, the Drosophila embryo has been used to explore in vivo microtubule dynamics during cellular division10,11,12. The use of microinjection to deliver small molecules and molecular probes provides temporal control that makes Drosophila embryonic development particularly powerful for probing cellular processes in vivo13,14. However, generating a significant number of experimental repeats with a traditional imaging approach is extremely labor intensive and has limited use in high-throughput imaging screens and quantitative analysis. Our approach makes use of a high content analyzer typically reserved for single cell analysis and adapts in vivo imaging analysis in the syncytium of the early Drosophila embryo.
This protocol was used to collect, stage, and microinject Drosophila embryos to explore the mechanisms that drive morphological changes in the Endoplasmic Reticulum (ER) during mitosis15. Although many studies have outlined the dynamic nature of the ER during the cell cycle16,17,18,19, it has been difficult to compare the effects of multiple perturbations across time on ER morphology. To identify the components that drive changes in ER morphology across cell division, the methods listed in this protocol were used to probe the role of microtubules and other cytoskeletal factors on mitotic ER reorganization using the cortical syncytial division in the early Drosophila embryo. Taken together, the availability of genetic perturbation within the Drosophila community, the ability to microinject drugs and molecular probes at precise times during development, and the inexpensive cost of working with Drosophila makes this approach highly amenable to high-throughput image-based screening. The methods described here are applicable for studying a wide variety of cellular and developmental processes in vivo using the Drosophila embryo20. Specifically, this protocol is well adapted to study biological events that occur over an extended period and within 100 microns of the Drosophila embryo cortex.
Protocol
NOTE: Refer to Table 1 for recipes of media and solution.
1. Setting up and maintaining an embryo collection cage for live analysis21
- Populate a fresh embryo collection cage with fresh flies.
NOTE: While commercially available collection cages are recommended, collection cages can be easily constructed from empty fly bottles21.- Discard or transfer adult flies from bottles when many pupae begin darkening at stage p14; at 20 °C this occurs approximately 14 days after the eggs have been laid22.
- Allow pupae to hatch and populate the bottles for 12-24 h.
- Transfer fresh flies into a small collection cage using a small funnel over the collection cage opening to prevent flies from escaping. Combine up to 3 bottles to produce a densely populated cage containing 200 to 500 flies.
- Transfer flies into the collection cage by tapping a fly bottle against a hard surface repeatedly (to stun the flies). Then open and invert the fly bottle over the collection cage funnel. Tap the open bottle onto the collection cage a few times to drop flies into the collection cage.
- Close the open fly bottle by quickly setting it down onto its plug. While transferring the open fly bottle from the funnel to its plug, ensure that the bottle opening points downward as flies tend to crawl up away from the source of the tapping.
- Press the funnel against the fly cage opening and tap the cage repeatedly on a hard surface to stun the flies inside. Immediately remove the funnel and secure a fresh grape plate to the fly cage opening. Use labeling tape to secure the grape plate. Repeat this process to combine up to 3 bottles to produce a densely populated collection cage.
NOTE: Alternatively, flies can be anesthetize using CO2 or other fly sleeping agents; however, this will increase the acclimation period in the step 1.2.
- Acclimate the new collection cage for 24-48 h. If CO2 or any other sleeping agents were used, acclimate the collection cage for at least 48 h.
- Replace the grape plate in the collection cage with a fresh one containing yeast paste twice a day, 8-10 h apart.
- Setup a fresh collection cage every 5 days.
2. Collecting and preparing embryos for imaging
- Obtaining embryos with synchronized development
- Exchange the grape plate on the collection cage with a fresh one containing a dab of yeast paste at the center. Allow flies to lay embryos for 1 h. Toss the old grape plate.
NOTE: Flies can fertilize embryos prior to laying them. The first collection plate will contain many of these predeveloped embryos. Allowing flies to dump old embryos first will improve developmental synchronization between embryos. - Exchange the grape plate for a fresh one containing a dab of yeast paste at the center and let flies lay embryos for 10 min. Toss the old grape plate.
NOTE: To avoid slowing down embryonic development on cold plates, warm the grape plate and yeast paste to room temperature prior to use. - Exchange the grape plate with a fresh one (without yeast paste) and save the old grape plate, it has the embryos for imaging. If necessary, continue collecting embryos from this cage by repeating step 2.1.2 and 2.1.3.
- Exchange the grape plate on the collection cage with a fresh one containing a dab of yeast paste at the center. Allow flies to lay embryos for 1 h. Toss the old grape plate.
- Age collect embryos for 30-45 min to image syncytial divisions23.
NOTE: To image the syncytial blastoderm stage in embryonic development, the embryos must be mounted and on the microscope within 90 min of collection. When attempting this protocol for the first time, age embryos 10-20 min in step 2.2 to provide extra time to set up between steps. - Dechorionate embryos with 50% bleach in DI water.
- Transfer aged embryos from step 2.2 into a 140 nm sieve by filling the grape plate with DI water, gently dislodging the embryos with a paint brush, and pouring the water with embryos into the 140 nm sieve (Figure 1A).
NOTE: This can be done over a sink or 1,000 mL beaker. - Vigorously rinse embryos with a DI water using a squirt bottle. Dry embryos by blotting the sieve on a dry paper towel. Blot the sieve until it stops leaving water marks on the paper towel (Figure 1B).
NOTE: Remove all yeast paste from embryos as it can render bleach treatment ineffective in the next step. - Fill an empty 10 cm plate with freshly prepared 50% bleach diluted with DI water (1:1 bleach: DI water). Dip the sieve into 50% bleach for 2 min 45 s. Start a stopwatch as soon as the sieve touches the 50% bleach. Blot the sieve in and out of the 50% bleach 3-4x in the first 15 s to breakup embryo clumps (Figure 1C).
- Vigorously rinse embryos with DI water using a squirt bottle. Dry the embryos by blotting the sieve on a dry paper towel. Repeat vigorous rinse and blot drying process for 5x (Figure 1D).
NOTE: This is the most important step for removing left over chorion. Use a full DI squirt bottle and use half of the bottle in this step. - Transfer the blot dried embryos from the sieve to a 10 cm grape plates using a paint brush21 (Figure 1E)
NOTE: Manual dichlorination can be used as an alternative to bleach decorination24.
- Transfer aged embryos from step 2.2 into a 140 nm sieve by filling the grape plate with DI water, gently dislodging the embryos with a paint brush, and pouring the water with embryos into the 140 nm sieve (Figure 1A).
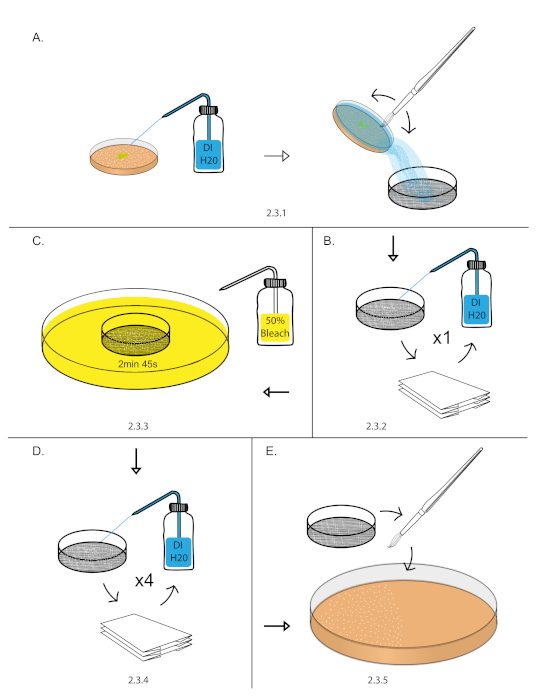
Figure 1: Embryo dechorionation. (A) Embryos were collected using a DI H20 squirt bottle, paint brush, and 140 nm sieve. (B) Embryos were rinsed vigorously using a DI H20 squirt bottle and blot dry over a paper towel. (C) Dechorionation of embryos was performed in 50% bleach for 2 min and 45 s. (D) Embryos were rinsed vigorously using a DI H20 squirt bottle and blot dry over a paper towel. This was repeated 4 times to remove excess chorion. (E) Embryos were then transferred to 10 cm grape plate using a paint brush. Please click here to view a larger version of this figure.
3. Mounting embryos to coverslips
- Under a stereomicroscope equipped with overhead goose neck lighting, organize two rows of 15-20 embryos on a clean section of grape plate using an egg picker tool. Align the embryos on their ventral side in the same orientation with respect to anterior and posteriors ends. To make egg pickers bend fine tip stainless steel tweezers to 45° angle. To review D/V embryos orientation see Campos-Ortega et al.20.
NOTE: It is important for the ventral surface to sit on the grape plate as the embryos will be pressed onto a cover slip in step 3.5, exposing the dorsal surface for imaging. Since the dorsal surface is much flatter than the ventral surface, imaging the dorsal side surface increases the number of nuclei present within a single Z plane. - Prepare 75 mm x 25 mm silicone spacer coverslip.
NOTE: This can be prepared ahead of time. Prepare several coverslips and store them in a slide box to keep them clean.- Trace the outline of a 75 mm x 25 mm coverslip onto a 1.6 mm thick silicone spacer sheet and cut out the silicone spacer using scissors (Figure 2A).
- Using a razor blade, cut out a 40 mm x 15 mm inset from the spacer (Figure 2A).
- Adhere the spacer onto the coverslip and streak 10 µL of heptane glue down the exposed coverslip glass inset (Figure 2A).
NOTE: Putting down and even streak will ensure imaging embryos on the same z plane.
- While the heptane glue from step 3.2.3 dries, cutout a rectangular section of grape plate containing the embryos from step 3.1.
- Use a razor blade to cut a rectangle (smaller than 40 mm x 15 mm) around the two rows of embryos. Do not remove the rectangle yet.
- Cut out a second rectangle next to the first. Remove the second rectangle using the straight edge side of a stainless-steel spatula.
- Carefully slide the straight edge of a stainless-steel spatula under the first rectangle and remove it (Figure 2B).
- Place the grape plate cutout with embryos on the external side of a 3.5 cm dish (Figure 2C).
- Under a stereomicroscope, lower a prepared coverslip over the embryos and gently press the two rows of embryos onto the glue (Figure 2D).
- If microinjecting embryos, skip to step 4.1. If not microinjecting continue to step 3.7.
- Cover embryos by filling the silicone spacer (halfway to the top) with 1:1 – halocarbon oil 700: halocarbon oil 27.
- Line the bottom edges of a 4-slide plate adapter with double sided tape to prevent the cover slip from moving but ensure not to place the tape in the path of the objective lens. This tape is critical. If the coverslip is only sitting on the adapter, it will move throughout the acquisition. Mount the cover slip on the 4 slide plate adapter.
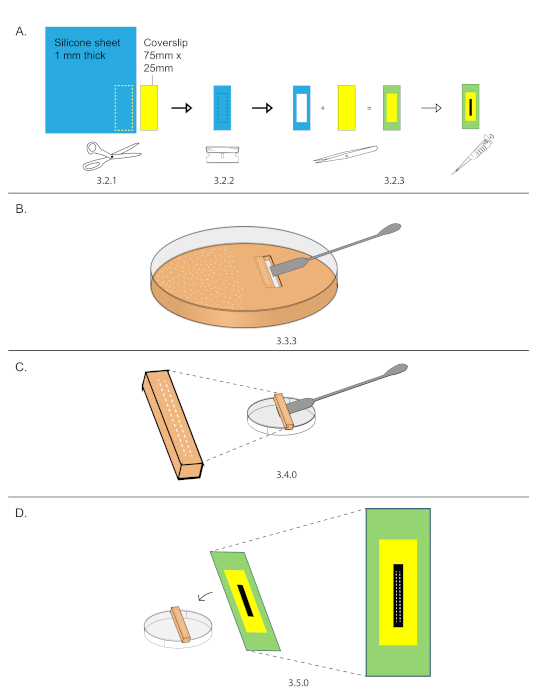
Figure 2: Coverslip preparation and mounting. (A) An outline of 75 mm x 25 mm coverslip was traced onto a 1.6 mm thick silicone sheet. Use scissors to cut out the outline. 40 mm x 15 mm inset were cut out from the silicone spacer and then mounted onto the coverslip. Streaked 15 µL down the center of the inset. (B) Two rows of 15-20 embryos were organized on a grape plate in an area smaller than 40 mm x 15 mm, then that area was cut out using a razor and stainless-steel spatula. (C) Grape plate cut out were placed on the top of an empty 3.5 cm dish. (D) Under a stereomicroscope, the coverslip from (A) was lowered and the embryos were stuck onto the streak of glue. Please click here to view a larger version of this figure.
4. Desiccating and microinjecting embryos
- Place the coverslip with embryos over a slide to prevent the bottom of the coverslip from becoming dirty during desiccation and microinjection.
NOTE: The cover slip will be imaged without a slide attached to it. Therefore, an inverted microscope is necessary for imaging. - Desiccate embryos for 5-10 min, depending on the temperature of the room. The embryos used for imaging in the representative results were desiccated for 7 min at 20 °C in a chamber containing a 1:1 layer of silica gel beads over a drying agent (Figure 3A).
- Immediately after desiccation, cover embryos with 1:1 – halocarbon oil 700: halocarbon oil 27. Fill the silicone spacer halfway to the top.
NOTE: It is important to mix a high viscosity halocarbon oil with a low viscosity halocarbon oil. Pure halocarbon oil 700 (high viscosity) slows down cell cycles progression 30 min post application. A 1:1 mixture of halocarbon oil 700 and halocarbon oil 27 fixed this artifact. - Load microinjection needles with injectant, then mount one to the microinjector and cut it open under pressure.
NOTE: Practice step 4.4 a few times before performing the full assay with any valuable reagents. This is the most difficult step in the microinjection process.- Load microinjection needles with 2 μL of injectant using an extended loading tip. Only one microinjection needle is required, but it is good to have spare loaded needles close by so prepare 2 or 3 needles.
NOTE: Refer to the Table of Materials for specifications on extended loading tips. Microinjection needles cannot be loaded without extended loading tips due to the unique nature of our microinjection technique, which relies on building air pressure inside of a sealed and loaded needle prior to opening its tip. - Mount a needle onto the microinjector and depress the plunger halfway to the tip. The goal is to increase air pressure inside of the glass capillary (Figure 4A).
NOTE: Use a plunger mineral oil based microinjector without mineral oil. Instead of building pressure with mineral oil, use the plunger to build air pressure. - Lower the glass needle onto a glass slide and cut the tip of the needle open using a new #9 razor. Cut the needle at a 45° angle to produce a sharp open tip. The injectant should flow down to the bottom of the tip without flowing out of the needle. Carefully add 20 μL of halocarbon oil over the tip of the needle and re-cut it open at 45°. The injectant should begin to flow rapidly.
- Immediately lower air pressure by retracting the plunger but ensure to maintain a continuous flow rate to prevent the needle from clogging with embryo membrane between injections. (Figure 4B).
NOTE: If the injectant is clear, view the injectant flow rate by lifting the microinjection needle in and out of halocarbon oil and observing the rate of formation for injectant droplets under a stereomicroscope. Adjust air pressure by depressing the plunger a few clicks at a time, then dip the microinjection tip in and out of halocarbon oil to view the rate of formation for new droplets post adjustment. - Use the micromanipulator to position the needle at the center of the field of view, then lift the needle using the micromanipulator and remove the glass slide from under it. The needle is now calibrated for injection.
- Load microinjection needles with 2 μL of injectant using an extended loading tip. Only one microinjection needle is required, but it is good to have spare loaded needles close by so prepare 2 or 3 needles.

Figure 3: Desiccation chamber diagram. (A) This is a diagram of a desiccation chamber. The chamber consists of a 1:1 layer of silica gel beads over a layer of drierite. To perform desiccation, embryos were placed on top of a single well plate lid. The desiccation chamber was closed 7 min. Please click here to view a larger version of this figure.
- Microinject embryos
- Place the embryos on the stereomicroscope stage under microinjection needle but do not lower the needle.
- Focus on embryos at 50x magnification.
- Lower the needle until it comes into the same focus plane as the embryos. Moving the slide with one hand and operating the microinjector with the other, inject one row of embryos.
- Raise the needle and rotate the slide 180° to expose the second row of embryos to the microinjection needle. Lower the needle and inject the second row of embryos.
NOTE: Try to do this in under 10 min. Leave some un-injected embryos to be used as controls.
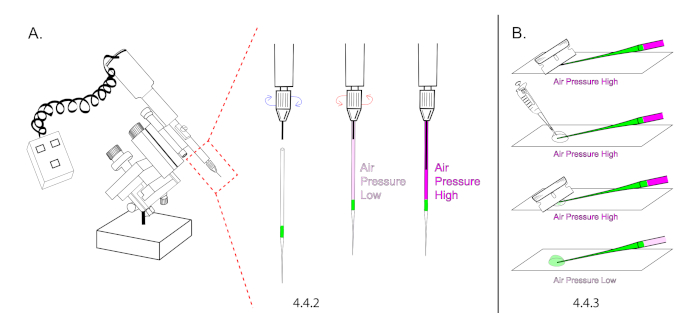
Figure 4: Microinjection needle preparation. (A) In this illustration, the intensity of magenta in needle is representative for air pressure. A loaded needle was mounted onto the microinjector and the plunger was extended 50% to increase air pressure inside of the needle. Make sure the plunger is maximally retracted prior to mounting. (B) The needle tip was cut under pressure and the plunger was retracted to decrease air pressure and the flow rate. Please click here to view a larger version of this figure.
5. Imaging on a high content analyzer
- Prior to running this assay create a plate map for a slide adapter. Use the plate holder editor found in Plate Map Manager menu to enter the dimension for a custom plate adapter.
NOTE: A custom plate map was made for the commercially available plate adapter (see Table of Materials). The plate map contains the following settings; Slide thickness: 1000 µm, Bottom height: 1.607 mm, bottom height variability: 100 µm, substrate material: glass, coverslip thickness: 150 µm, coverslip position: bottom – rows: 1 – spacing: 56.987 mm, columns: 4 – spacing 28.391, size and shape: rectangle, width 23.00 mm, height: 73.00 mm, offset to A1 21.29 mm – 42.74 mm) - Load the slide adapter on the microscope and use the Anteprima feature to tile a brightfield 10x image of the coverslip.
- Select an embryo from the top or bottom of a row, then select Objective and switch to 60x (NA 0.95). Determine an appropriate exposure for the desired fluorescence channel.
NOTE: Maintain the laser intensity under 25% for live cell imaging. - On the main dashboard, use the Anteprima feature to tile a fluorescent 60x image containing both rows of embryos.
- On the main dashboard, select the Binning drop down menu and set the binning to 1 x 1 for optimal resolution.
- Use the z Stack Setup tab to define parameters for z stacks.
- In the z Stack Setup tab, use the Up and Down arrows to find then define top and bottom z stack limits. Once the top and bottom limits are defined, select the Green Pencil Icon to save the z stack position. Note the z stack position as it will be used in step 5.6.5.
- In the z Stack Setup tab, use the z Step input box define spacing between z slices.
NOTE: The total number of slices is determined by modulating step 5.6.1 and 5.6.2. - On the main dashboard, right click on the Fluorescent Channel and use the Image Mode drop down menu to set the imaging mode to 3D.
- On the main dashboard, select the Laser Autofocus Box to disable autofocus.
- On the main dashboard, define the Initial Focus with the z-stack position from step 5.6.1 then select the Initial Focus Box to disable the “Refocus at each Time Point” feature.
- In the Fields tab, use the Field placement drop down menu to enable Custom Points, then use the Arrows to select points for imaging.
NOTE: The time interval between acquisitions (step 5.8) will determine how many custom points can be imaged simultaneously. - In the Time Series tab, select the Acquire Time Series box to enable the time series feature, then define the total number of time points.
NOTE: 6 custom points x 5 z slices x 80 stacks = 2,400 images, 2,400 images x 8.2 MB per image = 19.68 GB disk space. - On the main dashboard, use the Name Input Box to name the acquisition settings then select File and Save to save the acquisition settings.
- On the main dashboard, select the Scan button to run the acquisition.
6. Post image processing
- Drag and drop the xdce file into Fiji to open the image series as a multidimensional hyperstack.
- Save the hyperstack as a tif file.
- Create max intensity projections and save them as a tif files.
- Assemble a library of max intensity projections and build a pipeline for data extraction and analysis.
Representative Results
The approach presented in this protocol was used to examine the role of microtubules in Endoplasmic Reticulum (ER) reorganization during mitosis in the Drosophila embryo15. During mitosis, the structure of the ER displays a dramatic reorganization, however, the forces that drive these changes are poorly understood. Recently, a family of ER shaping proteins were shown to promote the formation of ER tubule25,26,27,28, which are known for their close proximity with microtubules28. To study the roll of microtubules on ER morphology, the methods provided in the protocol were used to generate data to quantitatively compare the effects of several microinjected drug treatments on ER reorganization during mitosis. Colchicine, which prevents new microtubule polymerization, was found to drastically reduce the reorganization of the ER during mitosis as shown in Figure 5 and Figure 6. Furthermore, the approach described here enabled the production of time laps imaging data for 32 embryos. Mean and max intensity measurements were produced from 12,800 regions of interest using a custom MATLAB script, which also generated descriptive and inferential statistics vital for making comparisons between drug treatments. Due to the availability of molecular probes and the ease of microinjections, the methods described here are easily adaptable to study a variety of biological processes via fluorescence intensity quantification.

Figure 5: Representative Images of Rtnl1-GFP and ReepB-GFP enrichment at the spindle poles during mitosis. (A) 60x field of view of ReepB-GFP during mitosis in cell cycle 10. The blue square represents the field of view used for panels B-E. Panel A is processed with a threshold binary filter. This filter was not applied to Panels B-E since it excludes data from pixels below the set threshold. (B,D) ReepB-GFP in control, and in colchicine. (C,E) Rtnl1-GFP in control, and in colchicine. This figure has been modified from Diaz et al.15. Please click here to view a larger version of this figure.
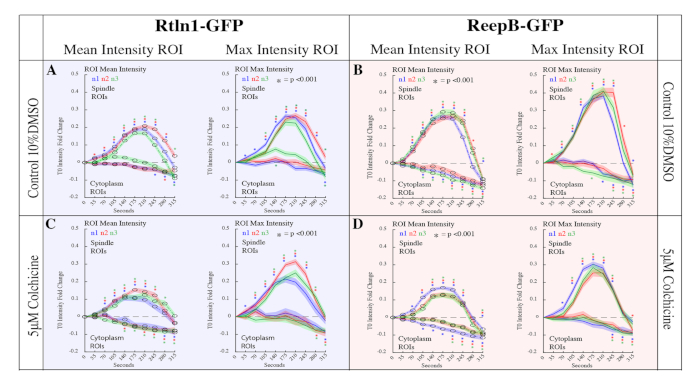
Figure 6: Quantitation of Rtnl1-GFP and ReepB-GFP enrichment at the spindle poles during mitosis. 20 spindles and 20 cytoplasm ROIs across 10 frames were analyzed for 32 embryos in the following conditions. (A) 10% DMSO control injections for Rtnl1-GFP. Control embryos showed a 0.1-fold increase in enrichment (p<0.001 for all samples at T210) compared to non-injected samples (p < 0.001 for all samples at T210). (B) ReepB-GFP embryos injected with 10% DMSO to serve as control. Control embryos showed a 0.1-fold increase in enrichment (p<0.001 for all samples at T210) compared to non-injected samples (Figure 1E; p < 0.001 for all samples at T210). (C) Rtnl1-GFP embryos injected with 5 μM Colchicine. Here we measured a reduction in Rtnl1-GFP enrichment to spindles (p < 0.001 for all samples at T210) compared to controls (A) (p < 0.001 for all samples at T210). (D) ReepB-GFP embryos injected with 5 μM Colchicine. We measured a reduction in ReepB-GFP enrichment to spindles (p<0.001 for all samples at T210) compared to controls (B) (p<0.001 for all samples at T210). This figure has been modified from Diaz et al.15. Please click here to view a larger version of this figure.
| 1. Yeast Paste21 | 2. Grape Plates21 | 3. Heptane Glue24 |
| 1.1 Add 7 grams of active dry yeast to a 50 ml conical. Add 9 milliliters of DI water and mix to even constancy with a metal spatula. | 2.1 Make solution a: Add 6 grams of agar to 140ml of DI water in a 250ml flask and autoclave it. | 3.1 Fill a glass scintillation vial with 50 inches of double-sided tape. |
| 1.1 Add 7 grams of active dry yeast to a 50 ml conical. Add 9 milliliters of DI water and mix to even constancy with a metal spatula. | 2.2 Make solution b: Mix 0.1 grams of methyl paraben in 2ml of ethanol, then add this solution to 60ml of grapefruit juice concentrate. | 3.2 Add 20 ml of heptane and seal glass scintillation vial with parafilm. Rotate overnight at room temperature. |
| 2.3 Immediately after autoclaving solution a, mix in solution b and pour mixture into 10 x 35mm perti dishes. (makes 35 grape plates) | 3.3 Remove plastic tape debris by transferring glue into 2 15 ml conical tubes and centrifuging at 3000 RPM for 15 minutes. | |
| 2.4 Allow grape plates to set at room temperature overnight with plate lids off. Cover plates with lids and store in 4C° after they set. | 3.4 Transfer glue supernatant into 1.5 ml microcentrifuge tubes for storage. |
Table 1: Recipes of media and solutions.
Discussion
The protocol described here is highly versatile and easily adaptable for studying biological events in a variety of stages in Drosophila embryonic development. This protocol is well suited for studying biological processes that occur over long periods of time. For example, studies of cellularization29,30 or the formation of heterochromatin domains in Drosophila31,32 could benefit from utilizing the methods described in this protocol since these processes occur over an extended period of time. In addition, sample size can be increased by decreasing magnification to study questions that require less optical resolution, such as the timing between syncytial divisions33 or duration of gastrulation34. Likewise, a 20x objective allows for two embryos to be imaged within one imaging plane. The methods in this protocol provide a dynamic platform to ask biological questions using embryonic development as a model system for quantitative analysis.
There are 3 critical steps in this protocol. Post dechorionation in 50% bleach it is imperative that embryos are rinsed and dried vigorously 5 times (2.3.4). This step removes any leftover small pieces of chorion. Leftover chorion will obstruct the field of view when imaging the embryo. When mounting the embryos onto the glass coverslip (3.5), it is important to press the embryos onto the glue with uniform pressure. This will place the embryos on the same z plane. When cutting open the microinjection needle, it is important to retract the plunger immediately to adjust the flow rate of the needle or you will lose all your injectant.
Our study was limited by two factors. The first factor was not having an automated segmentation process. We designed a manual segmentation script using MATLAB, however; with a genetically encoded spindle marker this process could be automated. In the future we would like to produce CNN-RFP21, Rtln1-GFP and CNN-RFP; ReepB-GFP transgenic lines to enable automated spindle segmentation. For more information regarding our MATLAB script and analysis pipeline, refer to supplemental materials of Diaz et al.15. Secondly, we were also limited by our acquisition interval of 35 s, which only allowed us to image 6 embryos simultaneously. A longer acquisition interval would allow us to image more embryos simultaneously. Although drug delivery throughput can be increase by replacing microinjection with a permeation step, we found this to be unnecessary since our sample size was already limited by acquisition interval.
While this protocol was used to image Drosophila embryonic development, the overall approach is adaptable for studying embryonic development in other multicellular model systems, such as C. elegans, Sea Urchins, and Xenopus. Embryos are extremely useful for quantitative analysis since they are easily synchronized via collection or fertilization. In addition, embryos don’t move or swim away from a field of view, enabling the use of a simple multipoint acquisition capable software to produce multiple time lapse videos for quantitative analysis. Although we used a high content analyzer in our study, any inverted microscope system capable of multipoint acquisition can be paired with the methods referred to herein.
Similar results can be obtained using a multipoint acquisition capable inverted confocal microscope; however, the advantages of a high content analyzer emerge as one increases sample size to study questions that require less resolution in time. An obvious advantage to using a high content analyzer is the ability to image 4 separate slides simultaneously compared to 1 slide on traditional systems. With a full set of 4 slides and 40 embryos on each slide, 160 embryos can be imaged over several hours, so long as the acquisition interval is at least 15 min between frames. Another important advantage to using a high content analyzer is that the samples are completely sealed off from any external light sources, which is necessary for fluorescence intensity-based measurements. Lastly, the high content analyzer used in our study has a 5.5-megapixel CMOS camera which provides a generous 221.87 μm field of view at 60x magnification. This larger field of view enabled us to image half an embryo per acquisition point.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
UD, WFM and BR are supported by the Center for Cellular Construction, a National Science Foundation (NSF) Science and Technology Center, under grant agreement DBI-1548297. BR is also supported through an NSF CAREER award, 1553695.
Materials
| 10 x 35mm Micro Dish | VWR | Cat #: 10799-192 | N/A |
| 100% grape juice concentrate | Welch's, 100% Concentrate Grape Juice | N/A | Purchase at Grocery Store |
| 250ml Erlenmeyer Flasks, Narrow Mouth | VWR | Cat #: 10536-914 | N/A |
| 3" Non-Metallic Sieve No. 140 | Gilson Company | SV-165 #140 | N/A |
| 4 Slide Imaging Tray | Grace Biolabs | SKU: 400104 | N/A |
| ACROS Organics , n-Heptane, 99+%, for spectroscopy | Fisher Scientific | Cat #: AC411255000 | N/A |
| Acros Organics, Ethanol, 99.5%, ACS reagent, absolute, 200 proof | Fisher Scientific | Cat# : AC615095000 | N/A |
| Acros Organics, Methyl 4-hydroxybenzoate, 99% . (methyl paraben ) | Fisher Scientific | Cat#: AC126961000 | N/A |
| Cover Slip, Rectangular, 25 x 75mm, #1 Float Glass | Fisher Scientific | Cat #: 50-143-782 | N/A |
| Culture Well Silicone Sheet Material – 1.6mm thick | Grace Biolabs | SKU: 664273 | N/A |
| DWK Life Sciences Wheaton 20mL PET Liquid Scintillation Vials | Fisher Scientific | Cat #: 03-341-71A | N/A |
| Embryo Collection Cage-Mini | Genesee Scientific | Cat #: 59-105 | N/A |
| Eppendorf Microloader Pipette Tips | Fisher Scientific | Product Code: 10289651 | N/A |
| Falcon 15mL Conical Centrifuge | Fisher Scientific | Cat #: 05-527-90 | N/A |
| Falcon 50mL Conical Centrifuge Tubes | Fisher Scientific | Cat #: 14-432-22 | N/A |
| Fleischmann's, Active Dry Yeast | Fleischmann's | N/A | Purchase at Grocery Store |
| Grape Plates | REFFER TO RECIPE PREP | N/A | N/A |
| Halocarbon 27 Oil | Genesee Scientific | Cat #: 59-133 | N/A |
| Halocarbon 700 Oil | Genesee Scientific | Cat #: 59-131 | N/A |
| Heptane Glue | REFFER TO RECIPE PREP | N/A | N/A |
| Industrial Razor Blades (Surgical Carbon Steel) Single Edged No. 9 | VWR | Cat #: 55411-050 | N/A |
| Parafilm M Laboratory Sealing Film, 4 inch x 250 foot Roll | Fisher Scientific | Cat #: 50-998-944 | N/A |
| Scotch Double Sided Tape with Dispenser, Narrow Width, Engineered for Bonding, 1/2" x 7 yds., 3/Pack Caddy | Fisher Scientific | Cat #: NC0879005 | N/A |
| Silica gel beads, desiccant ~ 2 – 5 mm | Millipore Sigma | SKU: 1077351000 | N/A |
| Stainless Steel Spoon/Spatula | VWR | Cat #: 470149-044 | N/A |
| W.A. Hammond Drierite Indicating Absorbents | Fisher Scientific | Cat #: 23-116582 | N/A |
| Yeast Paste | REFFER TO RECIPE PREP | N/A | N/A |
| EQUIPMENT | |||
| Allegra X-12 Benchtop, Centrifuge | Beckman Coulter | N/A | You will need access to 15 ml conical centrifuge inserts. |
| Autoclave | N/A | N/A | Any autoclave will do fine. |
| Drummond Nanoject II Microinjector. | Drummond Scientific | N/A | This is a mineral oil based injection system. |
| GE Healthcare IN Cell Analyzer 6000. | GE healthcare | N/A | A fast confocal microscope capable of multipoint acquisition may be substituted. |
| Refrigerator (4C°) | N/A | N/A | Fly media and yeast paste must be stored at (4C°). |
| Zies Stemi 508 Stereo Microscope with Transillumination base 300 and overhead gooseneck lighting. | Zeiss Microscopy | N/A | The transillumination base and gooseneck lighting are necessary. |
Riferimenti
- Zanella, F., Lorens, J., Link, W. High content screening: seeing is believing. Trends in Biotechnology. 28 (5), 237-245 (2010).
- Jennings, B. Drosophila-a versatile model in biology & medicine. Materials Today. 14 (5), 190-195 (2011).
- Soares, C., et al. 2D and 3D-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression. PLoS One. 7 (5), 38147 (2010).
- Bickle, M. The beautiful cell: high-content screening in drug discovery. Analytical and Bioanalytical Chemistry. 398 (1), 219-226 (2010).
- Boutros, M., Heigwer, F., Laufer, C. Microscopy based high content screening. Cell. 163 (6), 1314-1325 (2015).
- Mandal, A., Pinter, K., Drerup, C. Analyzing Neuronal Mitochondria in-vivo Using Fluorescent Reporters in Zebrafish. Frontiers in Cell and Developmental Biology. 6 (144), 00144 (2018).
- Bellen, H., Tong, C., Tsuda, H. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nature Reviews Neuroscience. 11 (7), 514-522 (2010).
- Letsou, A., Bohmann, D. Small flies-big discoveries: nearly a century of Drosophila genetics and development. Developmental Dynamics: An Official Publication of the American Association of Anatomists. 232 (3), 526-528 (2005).
- Sullivan, W., Theurkauf, W. The cytoskeleton and morphogenesis of the early Drosophila embryo. Current Opinion in Cell Biology. 7 (1), 18-22 (1995).
- Schejter, E., Wieschaus, E. Functional elements of the cytoskeleton in the early Drosophila embryo. Annual Review of Cell Biology. 9 (1), 67-99 (1993).
- Kellogg, D., Mitchison, T., Alberts, B. Behavior of microtubules and actin filaments in living Drosophila embryos. Development. 103 (4), 675-686 (1988).
- Mavrakis, M., Rikhy, R., Lilly, M., Lippincott-Schwartz, J. Fluorescence imaging techniques for studying Drosophila embryo development. Current Protocols in Cell Biology. 39 (1), 4-18 (2008).
- Tram, U., Riggs, B., Koyama, C., Debec, A., Sullivan, W. Methods for the study of centrosomes in Drosophila during embryogenesis. Methods in Cell Biology. 67, 113-123 (2001).
- Voeltz, G., Prinz, W., Shibata, Y., Rist, J., Rapoport, T. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124 (3), 573-586 (2006).
- Diaz, U., Bergman, Z., Johnson, B., Edington, A., de Cruz, M., Marshall, W., Riggs, B. Microtubules are necessary for proper Reticulon localization during mitosis. PLoS One. 14 (12), 0226327 (2019).
- Shibata, Y., et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. Journal of Biological Chemistry. 283 (27), 18892-18904 (2008).
- Bergman, Z., et al. Spatial reorganization of the endoplasmic reticulum during mitosis relies on mitotic kinase cyclin A in the early Drosophila embryo. PloS One. 10 (2), 0117859 (2015).
- Lu, L., Ladinsky, M., Kirchhausen, T. Cisternal organization of the endoplasmic reticulum during mitosis. Molecular Biology of the Cell. 20 (15), 3471-3480 (2009).
- Puhka, M., Vihinen, H., Joensuu, M., Jokitalo, E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. The Journal of Cell Biology. 179 (5), 895-909 (2007).
- Campos-Ortega, J., Hartenstein, V. . The embryonic development of Drosophila melanogaster. , (2013).
- Sulivan, W., Ashburner, M., Hawley, R. . Drosophila Protocols. , (2000).
- Bainbridge, S., Bownes, M. Staging the metamorphosis of Drosophila melanogaster. Development. 66, 57-80 (1981).
- Bate, M., et al. Mitosis and Morphogenesis in the Drosophila Embryo. The Development of Drosophila melanogaster. , (1993).
- Uyen, T., Blake, R., Carol, K., Alain, D., William, S. Methods for the study of centrosomes in Drosophila during embryogenesis. Methods in Cell Biology. 67, 113-123 (2001).
- Voeltz, G., Prinz, W., Shibata, Y., Rist, J., Rapoport, T. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124 (3), 573-586 (2006).
- Zurek, N., Sparks, L., Voeltz, G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 12 (1), 28-41 (2011).
- Kumar, D., Golchoubian, B., Belevich, I., Jokitalo, E., Schlaitz, A. L. REEP3 and REEP4 determine the tubular morphology of the endoplasmic reticulum during mitosis. Molecular Biology of the Cell. 30 (12), 1377-1389 (2019).
- Park, S., Zhu, P., Parker, R., Blackstone, C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. The Journal of Clinical Investigation. 120 (4), 1097-1110 (2010).
- Loncar, D., Singer, S. Cell membrane formation during the cellularization of the syncytial blastoderm of Drosophila. Proceedings of the National Academy of Sciences U.S.A. 92 (6), 2199-2203 (1995).
- Mazumdar, A., Mazumdar, M. How one becomes many: Blastoderm cellularization in melanogaster. Bioessay. 24, 1012-1022 (2002).
- Yuan, K., O’Farrell, P. TALE-light imaging reveals maternally guided, H3K9me2/3-independent emergence of functional heterochromatin in Drosophila embryos. Genes & Development. 30 (5), 579-593 (2016).
- Walther, M., et al. Heterochromatin formation in Drosophila requires genome-wide histone deacetylation in cleavage chromatin before mid-blastula transition in early embryogenesis. Chromosoma. 129 (1), 83-98 (2020).
- McCleland, M., Shermoen, A., O’Farrell, P. DNA replication times the cell cycle and contributes to the mid-blastula transition in Drosophila embryos. Journal of Cell Biology. 187 (1), 7-14 (2009).
- Tram, U., Riggs, B., Sullivan, W. Cleavage and Gastrulation in Drosophila Embryos. Encyclopedia of Life Sciences. , (2002).

