Agarose-Based Model Ecosystem for Cultivating Methanotrophs in a Methane-Oxygen Counter Gradient
Summary
A protocol is described for preparing a simple model ecosystem that recreates the methane-oxygen counter gradient found in the natural habitat of aerobic methane-oxidizing bacteria, enabling the study of their physiology in a spatially resolved context. Modifications to common biochemical assays for use with the agarose-based model ecosystem are also described.
Abstract
Aerobic methane-oxidizing bacteria, known as methanotrophs, serve important roles in biogeochemical cycling. Methanotrophs occupy a specific environmental niche within methane-oxygen counter gradients found in soils and sediments, which influences their behavior on an individual and community level. However, conventional methods to study the physiology of these greenhouse gas-mitigating microorganisms often use homogeneous planktonic cultures, which do not accurately represent the spatial and chemical gradients found in the environment. This hinders scientists’ understanding of how these bacteria behave in situ. Here, a simple, inexpensive model ecosystem called the gradient syringe is described, which uses semi-solid agarose to recreate the steep methane-oxygen counter gradients characteristic of methanotrophs’ natural habitats. The gradient syringe allows for the cultivation of methanotrophic strains and the enrichment of mixed methane-oxidizing consortia from environmental samples, revealing phenotypes only visible in this spatially resolved context. This protocol also reports various biochemical assays that have been modified to be compatible with the semi-solid agarose matrix, which may be valuable to researchers culturing microorganisms within other agarose-based systems.
Introduction
Microorganisms living at an anoxic-oxic interface often serve important ecological roles1. One example is aerobic methane-oxidizing bacteria (methanotrophs), which exist in counter gradients of methane and oxygen in soils and sediments2. These microorganisms possess unique metabolic and physiological characteristics that enable them to exploit the gas gradients present in their environments and have been the subject of ongoing research for decades3,4,5. Currently, most published research about methanotrophs and methane-oxidizing communities is based on work with homogenous planktonic cultures that often fail to capture the spatial and chemical gradients that are inherent to their natural microbial habitats. This limitation hinders our understanding of microbial physiology and our ability to link genomic information to phenotypic traits.
This protocol reports a simple, laboratory-based model ecosystem that creates reproducible conditions for studying both specific methanotrophs, such as Methylomonas sp. strain LW13, and methane-oxidizing communities directly from environmental soil samples. Importantly, cultivation in the gradient syringe results in counter gradient-specific phenotypes that are not present in homogenous planktonic cultures6, highlighting the system's ability to unveil new aspects of methanotroph physiology. Inspired by previously published model ecosystems7,8,9, the gradient syringe is a simplified method that can be used to collect chemical and molecular information from microorganisms cultured using this approach.
The reported procedures for genetic, chemical, and molecular analyses have been modified to work reliably on microbial cultures grown within a semi-solid agarose matrix. These procedures may also be useful for analyzing bacteria grown in other semi-solid agarose-based systems, such as those used for bacterial soft agar swimming assays. Adapting these analyses to spatially resolved contexts may open new avenues for studying microbial life in more ecologically relevant environments.
Protocol
The details of the reagents and the equipment used in the study are listed in the Table of Materials.
1. Preparation and extrusion of gradient syringes
NOTE: Gradient syringe preparation should be performed using sterile technique.
- Use several colonies of Methylomonas sp. LW13 freshly grown on a plate to inoculate 6 mL of nitrate mineral salts (NMS) medium in an 18 mm x 150 mm glass tube. Seal the tube with a serum stopper and aluminum crimp seal, and add methane using a syringe to a final atmosphere of 50% (v/v) methane in air. Shake this planktonic liquid culture at 200 rpm at room temperature until turbid (about one day).
- Passage liquid cultures 1:10 into fresh media. Continue growing liquid cultures of methanotrophs to log-phase growth (OD600 of ~0.5) and adjust to an OD600 =1.0.
- Prepare syringes by removing the accompanying plunger and keeping them in a sterile container. Attach a sterile PTFE filter tip to the syringe and place it in a standard test tube rack with the tip facing down.
- For each 10 mL syringe, thoroughly mix 1 mL of cells from step 1.2 with 5 mL of NMS and 4 mL of molten agarose (0.5% m/v, cooled to 55 °C) in a sterile conical tube. These volumes can be scaled up to fill multiple syringes in parallel.
- Slowly pour or use a serological pipet to add the mixture to each syringe, up to the 8 mL marking. Allow agarose within syringes to solidify (~15 min), then cap with a sterile 20 mm rubber butyl stopper. Secure the stopper to the syringe using lab tape and label it with syringe contents.
- To add methane to the syringe headspace, fill a large (60 mL) syringe with 100% CH4 and attach a PTFE filter tip (0.2 µm, 25 mm) connected to a sterile needle (23 G). Pierce the rubber stopper with the large syringe and pierce a second sterile needle through the stopper to create a gas outlet.
- Depress the plunger on the large syringe to allow 20 mL of 100% CH4 to flush through the headspace, taking care to remove the outlet needle when there is 1-2 mL of CH4 left in the large syringe to prevent oxygen backflow through the outlet needle.
- Incubate the syringes at 18 °C, repeating steps 1.6 and 1.7 daily to replenish the methane.
- To extrude agarose, replace the PTFE filter tip with a sterile 23 G needle and replace the rubber stopper with the supplied syringe plunger. Slowly depress the plunger to dispense 1 mL increments into separate sterile 1.5 mL microcentrifuge tubes.
2. Determining counter gradient gas concentrations
- Measuring the dissolved oxygen gradient
- Use a razor blade to slice through the width of an agarose-filled syringe (prepared following steps 1.1-1.8) close to the PTFE filter. Fasten the opened syringe to a syringe pump oriented towards a Clark-type microelectrode, with the open end facing the tip of the electrode.
- Adjust the settings of the syringe pump to move the syringe towards the microelectrode at a rate of 1 mL/min (0.6 cm/min); begin recording the dissolved oxygen measurements on the Unisense Logger software as soon as the syringe pump begins moving.
- Measuring the methane gradient
- Immediately before extrusion, replace the syringe filter tip with a one-way stopcock connected to a 23 G needle, and quickly swap the rubber stopper with a syringe plunger. Add eight 1 mL agarose aliquots to separate evacuated 12 mL gas-tight vials and allow the samples to equilibrate at room temperature for 1 h.
- Equilibrate sample vials to atmospheric pressure by cracking open and immediately resealing vials or piercing and quickly removing a needle. Inject 500 µL of vial headspace into a gas chromatograph with flame ionization detection (GC-FID) using a gas-tight syringe. Create a calibration curve derived from CH4 standards to convert peak area (pA*min) to µmol/L.
3. Counting cells in the gradient syringe
- Flow cytometry
- Extrude 1 mL of agarose segments from gradient syringes inoculated with either wild-type or mutant LW13 as outlined in step 1.9. Also, prepare and extrude agarose from a cell-free, sterile syringe as a negative control.
- Add 0.75 mL of 0.85% (m/v) NaCl in water to all extruded agarose samples and homogenize by vortexing. Further, dilute samples 1:10 by transferring 100 µL into a new microcentrifuge tube and adding 900 µL of the salt solution.
- Add 3 µL of a 1:1 mixture of SYTO9 and propidium iodide stains, then incubate in the dark at room temperature for 15 min. To determine the cells per mL of agarose, sonicate the microsphere counting bead suspension in a water bath for 5 min. Then, add 10 µL of the suspension to each sample before flow cytometry analysis.
- Analyze samples with a flow cytometer10,11 with the following parameters: triggering on green fluorescence, flow rate of 10 µL/s, and particle analysis rate below 1,000 particles/s.
- Compare SSC vs. FITC dot plots between cell-free control samples and inoculated agarose samples to draw "bacterial event" voltage gates that exclude background agarose particles. Additionally, draw voltage gates for the microsphere counting beads, which should be consistent between samples.
- To determine the concentration of cells in each agarose segment within the gradient syringe, use the following equation, noting that the dilution factor for the above protocol is 17.7275 and that 10-6 mL is the volume of one microsphere bead.
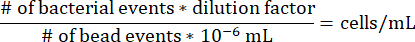
- Counting colony-forming units within the gradient syringe
- Extrude 1 mL of agarose segments into separate, sterile 2 mL microcentrifuge tubes, add 800 µL of NMS, and vortex for 10 s to aid in pipetting.
- Prepare a sterile 96-well plate by adding 180 µL of NMS to each well. Add 20 µL of diluted agarose samples to each well in the first column and pipet to mix.
- Using a multi-channel pipette, serially dilute samples tenfold by transferring 20 µL from the first row of wells into the second row of wells and pipetting 10 times to mix. Continue this process until the last row of the plate.
- Label square grid plates containing NMS agar or media of choice. Using a multi-channel pipette, spot 5 µL from a column of the 96-well plate onto the agar plate. Depending on the size of the agar plate, multiple columns can be spotted on the same plate.
- Incubate plates under 40% methane in air and grow at 18 °C. Count bacterial colonies after 2-3 days and determine the colony-forming units per milliliter (CFU/mL).
4. Biomolecule detection assays
- Polysaccharide assay
- Extrude 1 mL of agarose segments into separate 2 mL microcentrifuge tubes and mix with 1 mL of a 1% (m/v) Na2CO3 solution in water. Heat samples to 80 °C for 30 min with vortexing every 5-10 min, followed by centrifugation at 4,000 x g at 4 °C for 20 min.
- Collect the supernatant combined with three volumes of 100% ethanol and incubate at 20 °C for at least 2 h (or overnight).
- Collect the ethanol-precipitated polysaccharides by centrifugation at 16,100 x g at 4 °C for 30 min. Remove the supernatant and air-dry the pellet. Resuspend the pellet in 100 µL of deionized water.
- Measure the relative polysaccharide content of each agarose segment using a phenol-sulfuric acid colorimetric assay12,13. Combine 50 µL of the resuspended extract with 150 µL of concentrated sulfuric acid and 30 µL of 5% (v/v) phenol in water in a clear 96-well plate.
- Measure the absorbance at 490 nm using a microplate reader and calculate the relative polysaccharide content of each agarose segment as a percentage of the absorbance of the agarose segment closest to the PTFE filter.
- Protein assay
- Extrude agarose into separate 1.5 mL microcentrifuge tubes and transfer 100 µL of each aliquot to glass test tubes.
- Determine the total protein concentration using the test tube protocol of a BCA protein assay kit. Prepare albumin BSA standards with agarose extruded from a sterile syringe as the diluent.
- Extracellular DNA assay
- Extrude agarose into separate 1.5 mL microcentrifuge tubes and transfer 20 µL of each to 0.2 mL microcentrifuge tubes.
- Measure DNA concentrations using the commercially available 1x dsDNA high-sensitivity assay kit following the manufacturer's protocol.
5. RNA extraction
- Prepare the extraction buffer by combining the following in 800 mL of RNase-free water: 2.0 g of CTAB, 2.0 g of polyvinylpyrrolidone (PVP 40), 81.8 g of NaCl, 100 mM of Tris-HCl (pH 8.0), and 20 mM of EDTA. Bring up the volume to 1 L and autoclave; store at 4 °C.
- Aliquot the prepared extraction buffer and add 1% (v/v) final concentration of beta-mercaptoethanol just before use. Warm the buffer to 65 °C using a water bath or heat block.
- Divide each gradient syringe into 1 mL sections by extruding agarose into separate RNase-free 2 mL microcentrifuge tubes following the procedure in step 1.9.
- Centrifuge samples at 21,000 x g, 4 °C, for 15 min and discard the supernatant, keeping samples on ice.
- Add 600 µL of pre-warmed extraction buffer to each pelleted 1 mL of extruded agarose. Add approximately 200 µL of zirconia/silica beads and homogenize samples for 3 min at 30 Hz/s using a bead beater, pausing halfway to place samples on ice for 2 min.
- Centrifuge the samples at 15,000 x g, 4 °C, for 2 min to reduce foaming. Extract samples by adding 600 µL of chloroform: isoamyl alcohol (24:1) to tubes and vortexing for 10 s.
- Centrifuge the samples at 15,000 x g, 4 °C, for 8 min. Carefully transfer the upper aqueous phase to a new RNase-free microcentrifuge tube and add 600 µL of chloroform: isoamyl alcohol (24:1) to the transferred upper phase and vortex for 10 s.
- Centrifuge the samples at 15,000 x g, 4 °C, for 8 min and transfer the new upper aqueous phase to a new RNase-free microcentrifuge tube. Add an equal volume of isopropanol to the transferred upper phase and incubate samples for several hours at -20 °C. Optional: samples can be left at -20 °C overnight.
- Collect RNA-containing precipitates by centrifuging samples at 16,100 x g, 4 °C for 30 min.
- Discard the supernatant and wash the pellet with 300 µL of cold 75% (v/v) ethanol made with RNase-free water and centrifuge at 16,100 x g, 4 °C, for 5 min.
- Wash the pellets again following step 5.10.
- After removing the ethanol supernatant, let the pellets air dry for 15 min. Dissolve pellets in 100 µL of RNase-free water.
- Treat samples with DNase I at 37 °C for 30 min following the manufacturer's protocol.
- Inactivate DNase I by adding 300 µL of acid phenol: chloroform: IAA (125:24:1, pH 4.5). Vortex for 10 s and incubate at room temperature for 5 min.
- Centrifuge at 16,100 x g, 4 °C, for 5 min and keep the upper aqueous phase by transferring it to a new RNase-free microcentrifuge tube.
- Optional: Pool RNA from the same segment across all replicate syringes by combining the upper phases into a conical tube.
- Add 1 volume of isopropanol equal to the volume of the RNA-containing upper phase and add 7.5 M of LiCl to a final concentration of 0.8 M, inverting several times to mix.
- Incubate the samples for several hours at -20 °C. Optional: samples can be left at -20 °C overnight.
- Centrifuge the samples at 16,100 x g, 4 °C, for 15 min and carefully discard the supernatant. Wash the RNA-containing pellet twice by adding cold 70% (v/v) ethanol in RNase-free water, centrifuging at 16,100 x g, 4 °C, for 5 min, and removing the supernatant.
- Allow the pellets to air dry at room temperature for 10 min and resuspend in 50 µL of RNase-free water. Store the RNA samples at -80 °C.
- Optional, depending on the RNA quality: Samples can be re-purified using an RNA purification kit to remove small (<200 nt) RNAs.
- To confirm RNA samples do not contain residual DNA, use 1 µL of purified RNA as the template for PCR amplification using universal bacterial 16S rRNA gene primers 27F/1492R. Include two additional PCR reactions containing 1 µL of either genomic DNA (diluted 1:100) or nuclease-free water to serve as the positive and negative controls, respectively.
- Run the products on a 1% agarose TBE gel using gel electrophoresis14.
NOTE: RNA samples with no DNA contamination should result in the absence of a band in the sample lane. If RNA samples show DNA contamination, reprocess the samples starting from step 5.13. RNA is ready for downstream analysis and can optionally be analyzed to quantify its integrity by measuring the RNA Integrity Number (RIN).
Representative Results
Here, the gradient syringe model ecosystem was used to cultivate a single strain (the methanotroph Methylomonas sp. strain LW13) (Figure 1A)6, but it can also be used to enrich for a methane-oxidizing microbial community by direct soil inoculation (Figure 1B). The presence of a methane-oxygen counter gradient was validated by measuring the concentration of methane and oxygen across cell-free and inoculated syringes (Figure 1C). For LW13-inoculated gradient syringes, a counter gradient formed within one day of flushing the syringe, which steepened over three days of incubation. Over the same time period, a horizontal band formed at the same depth at which both gas substrates reached their lowest concentrations (Figure 1A). The steep gas gradient and depletion of methane and oxygen past the depth of the horizontal band showed that LW13 aerobically metabolized methane and produced a phenotype not observed in homogenous planktonic culture. This phenotype is also produced by other methanotrophic bacteria isolated from the same environmental sample as LW136. Strain-dependent variation in the timing and depth of the horizontal band development among different methanotrophic strains suggested that the horizontal band was affected by the specific behavior of each microbe when cultivated in a spatially resolved context6.
The number of cells in the entire agarose plug was measured using flow cytometry and colony counts (CFU/mL) (Figure 2A). This method was used to compare the cell distribution and survival of wild-type LW13 to a mutant strain of LW13 containing a deletion in the fucose 4-O-acetyltransferase (OAT) gene, which was previously shown to influence horizontal band development6. The ΔOAT mutant of LW13 had lower overall growth in the gradient syringe over 6 days compared to the wild-type, an effect that was not observed in homogenous planktonic cultures of the same strains6. The mutant strain did not form the same distinct horizontal band as the LW13 wild type when cultured in the gradient syringe (Figure 2B). Cell numbers and horizontal band appearance were restored to levels similar to the wild type upon gene complementation in the mutant strain. These results demonstrate that the gradient syringe can be used to link genes to specific phenotypes only present in the methane-oxygen counter gradient.
A variety of genetic, chemical, and molecular techniques were adapted for use with bacteria grown within a semi-solid agarose matrix. The gradient syringe model ecosystem can be readily used for standard biomolecular quantification assays with the inclusion of uninoculated agarose as the negative control. The concentration of three different biomolecules commonly found in extracellular polymeric substances and biofilms was measured: polysaccharides, protein, and extracellular DNA15 (Figure 3). In LW13-inoculated syringe segments, the horizontal band had significantly more polysaccharides than other segments, with no significant increase in protein or extracellular DNA.
RNA-seq was used to measure transcriptional differences in LW13 growing at different depths of the syringe. Robust RNA extraction was achieved using a CTAB-based extraction buffer followed by conventional phenol: chloroform extraction and precipitation steps. The results from the RNA-seq analysis were later used to identify genes implicated in the production of the horizontal polysaccharide band. These results indicate that the semi-solid agarose essential for the creation of a spatially resolved model ecosystem does not prevent further biochemical analyses that are generally reserved for planktonic and plate-based cultures.

Figure 1: The gradient syringe model ecosystem. (A) The gradient syringe inoculated with the methanotroph Methylomonas sp. LW13. A distinct horizontal band (arrowhead) develops within two days of flushing the syringe with 100% methane. (B) Close-up photos of gradient syringes inoculated with soil diluted 10-1 and 10-4 and incubated for two weeks. Gradient syringes containing more dilute soil resulted in spherical colonies throughout the agarose, whereas more concentrated soil inocula resulted in a distinct band. (C) Characterization of the methane-oxygen counter gradient in LW13-inoculated and sterile gradient syringes after three days of incubation. The gray bar indicates the range of depths at which the polysaccharide band was located; data show the mean ± SD of three independent experiments with three technical replicates each. Panels (A) and (C) were modified from Beals et al.6. Please click here to view a larger version of this figure.
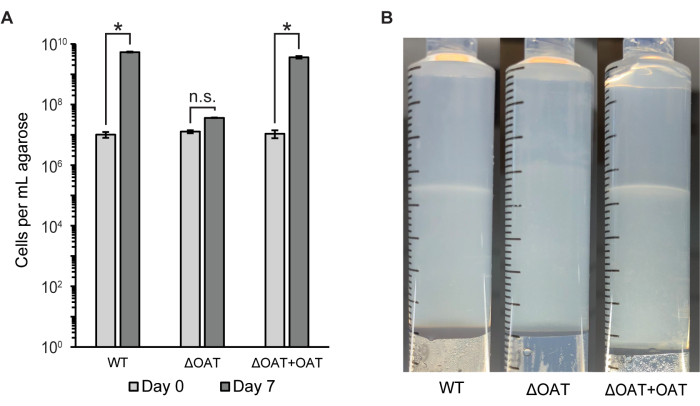
Figure 2: Quantification of LW13 wild type and mutant after incubation in the gradient syringe. (A) The total number of LW13 cells per mL extruded agarose recovered from gradient syringes on Day 0 and Day 7 measured by flow cytometry. ΔOAT contains a deletion of the fucose 4-O-acetyltransferase gene, which was found to be highly expressed in cells located at the depth of the polysaccharide band. ΔOAT+OAT contains the OAT gene inserted at a distal location of the ΔOAT genome. *, significantly different (two-tailed heteroscedastic t-test, α = 0.05); n.s., not significantly different. Data show the mean ± SD of three independent experiments with two technical replicates each. (B) Horizontal band development in gradient syringes inoculated with LW13 wild type, ΔOAT, or ΔOAT+OAT after seven days of incubation. This figure was modified from Beals et al.6. Please click here to view a larger version of this figure.
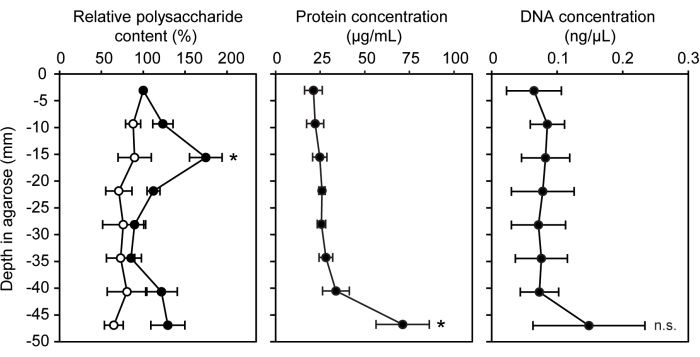
Figure 3: Quantification of biomolecules at increasing depths of the gradient syringe. Relative polysaccharide content (%), protein concentration (µg/mL), and DNA concentration (ng/µL) in eight sections of LW13-inoculated gradient syringes incubated for seven days (filled circles; open circles show values from sterile gradient syringes). Data show the mean ± SD of three independent experiments with three technical replicates each. For relative polysaccharide content, * indicates a significant difference from sterile control at equivalent depth (two-tailed heteroscedastic t-test, α = 0.05). For protein and DNA concentrations, * indicates a significant difference from the section containing the horizontal band (one-way ANOVA with Tukey-Kramer post hoc analysis); n.s., not significant. The figure was modified from Beals et al.6. Please click here to view a larger version of this figure.
Discussion
Methods for methanotroph cultivation
Methanotrophs have been studied for decades to understand their physiology, their individual and community behavior in the natural environment, and their potential for methane mitigation in industrial applications. Throughout these studies, much of the research conducted has been performed using homogenous planktonic cultures where spatial context is lost. The gradient syringe model ecosystem was developed to replicate the methane-oxygen counter gradient characteristic of natural methanotroph habitats in the lab, allowing researchers to study methanotrophs growing in an environment that more closely resembles where these organisms evolved.
Over the past 30 years, researchers have recreated the methane-oxygen counter gradient in the lab using a variety of methods, often with the primary goal of isolating and classifying methanotrophs from mixed methane-oxidizing consortia. These methods can be divided into two approaches, both involving the use of opposing chambers of methane and oxygen: suspending relatively undisturbed soil on a membrane16,17,18, or inoculating small amounts of soil or pure bacterial culture into a minimal medium in agarose7,8,19. The gradient syringe method described here combines the syringe-based approach of Dedysh and coworkers9 with the cultivation of methanotrophs from previous work by Amaral and Knowles8, and Schink and coworkers7. The latter of these methods laid the foundation for cultivating methanotrophs in a methane-oxygen counter gradient and used a continuous flow of methane and oxygen on either side of the agarose plug. While this provides a more constant environment, this approach adds complexity to the experimental setup and necessitates dedicated gas sources.
In contrast, the gradient syringe described here relies on daily flushing of the syringe to provide fresh methane, a process that takes less than a minute per syringe, while providing continuous access to atmospheric oxygen through a sterile PTFE filter tip. This simpler method may enable wider adoption of this model ecosystem for studying methanotrophs in a spatially resolved context. The described protocol also details chemical and molecular-level analyses that can be performed directly on bacteria incubated in the semi-solid agarose. As a result, bacteria do not need to be excised and cultured outside the agarose matrix before analysis, preserving the gas gradient conditions at the time of sampling.
Remarks on the protocol
Because the bacteria are cultured within a polypropylene volumetric syringe, researchers can use the accompanying syringe plunger to accurately and reproducibly segment the agarose plug while maintaining the spatial integrity of the agarose matrix that still remains in the syringe barrel. Without the air-tight design inherent to the syringe, agarose plugs would need to be removed from the syringe barrel and sliced, introducing uncertainty in the volume of agarose segments, and releasing unquantifiable amounts of methane and dissolved oxygen into the atmosphere. Agarose extrusion through a sterile needle simplifies sample preparation and helps homogenize extruded segments without shearing bacterial cells. This method allows researchers to divide each inoculated gradient syringe into at least eight agarose segments and perform parallel experiments on methanotrophs growing in a range of oxygen and methane concentrations.
In optimizing RNA extraction from high-polysaccharide content agarose, it was found that common reagents like guanidium thiocyanate and TRIzol led to agarose gelation, which obstructed purification columns and resisted pelleting by centrifugation. Low RNA yields and quality were also a concern as large polysaccharide molecules can trap nucleic acids while small polysaccharides can co-precipitate with RNA20. Instead, an extraction buffer containing the cationic surfactant CTAB was used, which solubilizes lipid membranes20; and NaCl, which prevents CTAB-nucleic acid complexes from forming and allows nucleic acids to precipitate but keeps polysaccharides in solution21. RNases were denatured by the inclusion of β-mercaptoethanol in the CTAB buffer. For the RNA-seq experiment, an optional column-based purification step was included to exclude small (<200 nucleotides) RNAs before library preparation.
Limitations and considerations
While the NMS and agarose provide a minimal medium matrix for cultivating methanotrophic bacteria, the gradient syringe as described here only recreates the gas gradients of methanotrophic habitats, but not other gradients present in those environments such as trace metals22, salinity23, or other nutrients24. It is possible these gradients can be added to a similar system in the future. Additionally, the volume of the syringe (8 mL agarose) limits the total biomass per syringe, necessitating pooling multiple syringes for some analyses (as described in step 5.16). Although the handheld syringe conveniently segments the agarose into 1 mL aliquots, its size also limits the headspace to approximately 4 mL, limiting the amount of bulk methane that can be stored for the cultivated microbes. Since methane oxidation rates are proportional to the growth rate of aerobic methanotrophs25, daily replenishment of headspace methane is recommended. While this may still result in periods of methane limitation, these periods are reproducible in the laboratory and likely mimic situations found in natural environments.
While using the gradient syringe, the presence of the agarose polysaccharides necessitates some adjustments to the assays used to analyze methanotrophs grown in this system. For example, protocols requiring the transfer of small volumes of extruded agarose need multiple dilution steps with thorough homogenization between each dilution for accurate pipetting. Additionally, in cases such as the polysaccharide assay where the polysaccharides inherent to the agarose matrix will react with the sulfuric acid-phenol reagent, the inclusion of a sterile, cell-free agarose negative control is essential. Early attempts to mitigate these issues by including the agarose-hydrolyzing enzyme β-agarase were unsuccessful and introduced an unknown variable to the biological experiments. The use of multiple technical replicates, thorough dilution, homogenization, and the inclusion of controls can be used to mitigate most of the challenges inherent to the agarose matrix.
Applications
In addition to single-strain studies, the gradient syringe can support the co-culture of multiple strains, and soil can be used as the inoculum in place of pure bacterial culture. The simple design of the gradient syringe model ecosystem is amenable to the culture of other types of microorganisms that exist at the interface between anoxic and oxic environments by using a different gas substrate, such as H2 or CO, in place of methane. In summary, the use of a simple, spatially resolved model ecosystem allows researchers to study the unique physiology and metabolic adaptations of anoxic-oxic microorganisms and can be used to link genes with organismal phenotypes.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by startup funding from the University of Utah Department of Chemistry and NSF CAREER Award #2339190. We thank members of the Puri Lab for helpful discussions. We thank Rachel Hurrell (University of Utah) for initial guidance with the flow cytometry experiment.
Materials
| 1% Gas mix analytical standard | Supelco | 22561 | 1% each component in nitrogen: carbon monoxide, carbon dioxide, hydrogen, methane and oxygen |
| 100% Methane | Airgas | ME CP300 | chemically pure grade |
| 15 ppm Gas mix analytical standard | Supelco | 23470-U | 15 ppm each component in nitrogen: methane, ethane, ethylene, acetylene, propane, propylene, propyne, and n-butane |
| 1x Nitrate mineral salts | see CAS numbers below | Dissolve the following in Mili-Q water and autoclave: 0.2 g/L MgSO4·7H2O, 0.2 g/L CaCl2·6H2O, 1 g/L KNO3, and 30 μM LaCl3. Before use, add trace elements to a 1X final concentration and phosphate buffer (pH 6.8) to a final concentration of 5.8 mM. | |
| 23 G needle | BD Biosciences | 305194 | sterile, Luer-Lok |
| 500x Trace elements | see CAS numbers below | Dissolve the following in Milli-Q water: 1.0 g/L Na2-EDTA, 2.0 g/L FeSO4·7H2O, 0.8 g/L ZnSO4·7H2O, 0.03 g/L MnCl2·4H2O, 0.03 g/L H3BO3, 0.2 g/L CoCl2·6H2O, 0.6 g/L CuCl2·2H2O, 0.02 g/L NiCl2·6H2O, and 0.05 g/L Na2MoO·2H2O. | |
| 96 Well plate | CELLTREAT | 229596 | sterile |
| Acid phenol:chloroform:IAA (125:24:1) | Invitrogen | AM9720 | pH 4.5 |
| Agarose | Fisher Scientific | BP160 | molecular biology grade, CAS 9012-36-6 |
| Aluminum crimp seals | VWR | 30618-460 | 20 mm |
| Bead beater | Qiagen | 9003240 | TissueLyser III |
| Butyl rubber stopper | Chemglass Life Science | 50-143-854 | 20 mm, blue |
| Chloroform:isoamyl alcohol (24:1) | Millipore Sigma | 25666 | BioUltra, for molecular biology |
| Clark-type O2 microelectrode | Unisense | OX-500 | |
| DEPC-treated water | Thermo Scientific | R0601 | |
| DNase I (Ambion) | Invitrogen | AM2222 | |
| Flow cytometer | Beckman Coulter | CytoFLEX | |
| Gas chromatograph (flame ionization detection) | Agilent | 6890N | |
| Gastight analytical syringe | Hamilton | 81220 | 1750 TLL |
| Gastight analytical syringe needle | Hamilton | 7729-07 | 22 G, metal hub needle, 2 in, point style 5 |
| Gas-tight vials | Labco | 938W | Exetainer vial: 12 mL, round bottom |
| Glass culture tubes | Bellco Glass | 2048-00150 | 18 x 150 mm |
| LiCl precipitation solution (7.5 M) | Invitrogen | AM9480 | |
| One-way stopcock | VWR | MFLX30600-00 | inlet port: female luer, outlet port: male luer lock |
| Petri dish, square | Fisher Scientific | FB0875711A | 100 x 100 mm |
| Phosphate buffer, 0.2 M (pH 6.8) | see CAS numbers below | Dissolve the following in Milli-Q water and autoclave: 12.24 g/L KH2PO4, 26.29 g/L Na2HPO4 · 7H2O | |
| Pierce BCA Protein Assay Kit | Thermo Scientific | 23225 | |
| PTFE syringe filter tip | Thermo Scientific | 03-050-469 | hydrophobic, pore size: 0.2 µm, diameter: 4 mm |
| Qubit 1x dsDNA High Sensitivity Assay Kit | Invitrogen | Q33230 | |
| Qubit 4 Fluorometer | Invitrogen | Q33238 | |
| RNA Clean & Concentrator-5 | Zymo Research | R1013 | |
| Serum stopper | Fisher Scientific | 03-340-302 | 20 mm |
| Syringe | BD Biosciences | 302995 | Luer-Lock, 10 mL, single use, sterile |
| Syringe pump | New Era Pump Systems Inc. | 1000-US | NE-1000 one channel programmable |
| SYTO9, propidium iodide, microspheres | Invitrogen | L34856 | LIVE/DEAD BacLight Bacterial Viability Kit |
| Zirconia/silica beads | BioSpec Products | 11079101z | 0.1 mm diameter |
| Chemical reagents | CAS number | ||
| CaCl2·6H2O | 7774-34-7 | ||
| CoCl2·6H2O | 7791-13-1 | ||
| Concentrated sulfuric acid | 7664-93-9 | ||
| CTAB, cetrimonium bromide | 57-09-0 | ||
| CuCl2·2H2O | 10125-13-0 | ||
| Ethanol | 64-17-5 | ||
| FeSO4·7H2O | 7782-63-0 | ||
| H3BO3 | 10043-35-3 | ||
| Isopropanol | 69-63-0 | ||
| KH2PO4 | 7778-77-0 | ||
| KNO3 | 7757-79-1 | ||
| LaCl3 | 10099-58-8 | ||
| MgSO4·7H2O | 10034-99-8 | ||
| MnCl2·4H2O | 13446-34-9 | ||
| Na2CO3, sodium carbonate | 497-19-8 | ||
| Na2-EDTA | 139-33-3 | ||
| Na2HPO4 · 7H2O | 7782-85-6 | ||
| Na2MoO·2H2O | 10102-40-6 | ||
| NaCl, sodium chloride | 7647-14-5 | ||
| NiCl2·6H2O | 7791-20-0 | ||
| Phenol (90% solution in water) | 108-95-2 | ||
| PVP40, polyvinylpyrrolidone | 9003-39-8 | ||
| Tris-HCl | 1185-53-1 | ||
| ZnSO4·7H2O | 7446-20-0 | ||
| β-Mercaptoethanol | 60-24-2 |
References
- Brune, A., Frenzel, P., Cypionka, H. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol Rev. 24 (5), 691-710 (2000).
- Auman, A. J., Stolyar, S., Costello, A. M., Lidstrom, M. E. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol. 66 (12), 5259-5266 (2000).
- Whittenbury, R., Phillips, K. C., Wilkinson, J. F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 61 (2), 205-218 (1970).
- Koo, C., Rosenzweig, A. Biochemistry of aerobic biological methane oxidation. Chem Soc Rev. 50 (5), 3424-3436 (2021).
- Strong, P. J., Xie, S., Clarke, W. P. Methane as a resource: Can the methanotrophs add value. Environ Sci Technol. 49 (7), 4001-4018 (2015).
- Beals, D. G., Puri, A. W. Linking methanotroph phenotypes to genotypes using a simple spatially resolved model ecosystem. ISME J. 18 (1), wrae060 (2024).
- Bussmann, I., Rahalkar, M., Schink, B. Cultivation of methanotrophic bacteria in opposing gradients of methane and oxygen. FEMS Microbiol. Ecol. 56 (3), 331-344 (2006).
- Amaral, J. A., Knowles, R. Growth of methanotrophs in methane and oxygen counter gradients. FEMS Microbiol Lett. 126 (3), 215-220 (1995).
- Danilova, O. V., et al. A new cell morphotype among methane oxidizers: a spiral-shaped obligately microaerophilic methanotroph from northern low-oxygen environments. ISME J. 10 (11), 2734-2743 (2016).
- Ou, F., McGoverin, C., Swift, S., Vanholsbeeck, F. Absolute bacterial cell enumeration using flow cytometry. J Appl Microbiol. 123 (2), 464-477 (2017).
- Krause, S. M. B., et al. Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc Natl Acad Sci USA. 114 (2), 358-363 (2017).
- Masuko, T., et al. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem. 339 (1), 69-72 (2005).
- Felz, S., Al-Zuhairy, S., Aarstad, O. A., van Loosdrecht, M. C. M., Lin, Y. M. Extraction of structural extracellular polymeric substances from aerobic granular sludge. J Vis Exp. (115), e54534 (2016).
- Lee, P. Y., Costumbrado, J., Hsu, C. Y., Kim, Y. H. Agarose gel electrophoresis for the separation of DNA fragments. J Vis Exp. (62), e3923 (2012).
- Costa, O. Y. A., Raaijmakers, J. M., Kuramae, E. E. Microbial extracellular polymeric substances: Ecological function and impact on soil aggregation. Front Microbiol. 9, 1636 (2018).
- Sinke, A. J. C., Cottaar, F. H. M., Buis, K., Keizer, P. Methane oxidation by methanotrophs and its effects on the phosphate flux over the sediment-water interface in a eutrophic lake. Microb Ecol. 24 (3), 259-269 (1992).
- Murase, J., Frenzel, P. A methane-driven microbial food web in a wetland rice soil. Environ Microbiol. 9 (12), 3025-3034 (2007).
- Reim, A., Lüke, C., Krause, S., Pratscher, J., Frenzel, P. One millimetre makes the difference: High-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic-anoxic interface in a flooded paddy soil. ISME J. 6 (11), 2128-2139 (2012).
- Rahalkar, M., Bussmann, I., Schink, B. Methylosoma difficile gen. nov., sp. nov., a novel methanotroph enriched by gradient cultivation from littoral sediment of Lake Constance. Int J Syst Evol Microbiol. 57 (5), 1073-1080 (2007).
- Wang, L., Stegemann, J. P. Extraction of high-quality RNA from polysaccharide matrices using cetlytrimethylammonium bromide. Biomaterials. 31 (7), 1612 (2010).
- Liyanage, N. M. N., Chandrasekara, B. C. H. W. M., Bandaranayake, P. C. G. A CTAB protocol for obtaining high-quality total RNA from cinnamon (Cinnamomum zeylanicum Blume). 3 Biotech. 11 (4), 201 (2021).
- Semrau, J. D., DiSpirito, A. A., Gu, W., Yoon, S. Metals and methanotrophy. Appl Environ Microbiol. 84 (6), e02289-e02317 (2018).
- Zhang, S., et al. Salinity significantly affects methane oxidation and methanotrophic community in Inner Mongolia lake sediments. Front Microbiol. 13, 1067017 (2023).
- Fox, A. L., Trefry, J. H. Nutrient fluxes from recent deposits of fine-grained, organic-rich sediments in a Florida estuary. Front Mar Sci. , (2023).
- He, L., et al. A methanotrophic bacterium to enable methane removal for climate mitigation. Proc Natl Acad Sci. 120 (35), e2310046120 (2023).

.
