3D Reconstruction and Analysis of Thin Subcellular Neuronal Structures using Focused-Ion Beam Scanning Electron Microscopy Data
Summary
A flexible methodological pipeline to identify, visualize, and quantify thin subcellular neuronal processes within focused ion beam scanning electron microscopy image volumes using user-friendly open-source software packages.
Abstract
Recent advances in scanning electron microscope technologies now permit the rapid three-dimensional (3D) analysis of ultrathin subcellular processes. Here, a methodological pipeline is presented to identify, visualize, and analyze thin neuronal processes, such as those that project into the presynaptic boutons of other neurons (termed ‘spinules’). Using freely available software packages, this protocol demonstrates how to use a decision tree to identify common neuronal subcellular structures using morphological criteria within focused ion beam scanning electron microscopy (FIB-SEM) image volumes, with particular attention on identifying a diversity of spinules projecting into presynaptic boutons. In particular, this protocol describes how to trace spinules within neuronal synapses to produce 3D reconstructions of these thin subcellular projections, their parent neurites, and postsynaptic partners. Additionally, the protocol includes a list of freely available open-source software programs for analyzing FIB-SEM data and offers tips (e.g., smoothing, lighting) toward improving 3D reconstructions for visualization and publication. This adaptable protocol offers an entry point into the rapid nanoscale analysis of subcellular structures within FIB-SEM image volumes.
Introduction
Investigations into the structure-function relationships of nanometer-thin subcellular components often benefit from 3D visualization and analysis1. However, serial section transmission electron microscopy studies have been temporally and spatially constrained by the necessity to use a diamond knife to cut and align hundreds to thousands of ≥40 nm serial ultrathin sections. These constraints have limited the ability to sample and effectively analyze thin (<40 nm in diameter) subcellular structures, and the necessity to become proficient at ultrathin serial sectioning has hampered the application of 3D structural analyses2,3. However, recent advancements in focused ion beam scanning electron microscopy (FIB-SEM) have revolutionized the speed and resolution of obtainable image volumes and now permit the quantitative analysis of thin subcellular structures such as smooth endoplasmic reticula4,5, neuronal synapses3,6, and synaptic vesicles7,8 at scale. In addition, wider use of FIB-SEM image volumes has accelerated the development of freely accessible FIB-SEM image volume repositories9 and 3D analysis software (e.g., Espina10, IMOD11, Neuromorph12, Reconstruct13, TrakEM14) that expand the reach of this technology and now enable investigations into the structure and function of fine subcellular structures.
One such nanoscale subcellular feature is the neuronal synaptic 'spinule.' Spinules are thin (~0.06-0.15 µm wide, 0.1-1 µm long), finger-like projections that emanate from one neuron and become encapsulated by the neurite (e.g., presynaptic bouton) of another neuron15,16. Spinules embedded within neuronal processes have been reported in the electron microscopy literature for almost 60 years17, and spinule-like protrusions are a conserved18,19,20 and ubiquitous21,22 feature of excitatory synapses. Nevertheless, despite the pervasiveness of synaptic spinules, their function(s) remain obscure, and there is a dearth of necessary data to explain their abundance and structural conservation. This lack of experimental characterization of spinules has mostly been due to the difficulty in quantitatively analyzing spinule prevalence and sizes. Their small dimensions are most suited to analyses using a previously unattainable z (depth) resolution (i.e., ≤15 nm).
Here, a FIB-SEM analysis pipeline is presented for identifying, visualizing, and analyzing thin (with cross sections ≤40 nm wide) subcellular structures that can serve as an entry point for FIB-SEM newcomers and experts alike. This protocol serves as a primer for identifying neuronal subcellular structures within a 3D FIB-SEM image volume, emphasizing how to use specific criteria to recognize and classify subtypes of spinules and synapses. Additionally, the protocol demonstrates how to import image volumes into a free 3D analysis software platform (Reconstruct), use this software to trace spinules within excitatory neuronal synapses, and produce 3D reconstructions of these subcellular projections, their parent neurites, and encapsulating presynaptic boutons. Lastly, the protocol shows how to use free, open-source 3D rending software (Blender) to smooth the 'skin' on 3D reconstructions for visualization and potential publication, detailing the advantages and potential pitfalls of this technique.
Protocol
1. Image volume data and subcellular object size: considerations and registration
- Obtain a quality FIB-SEM image volume of the area of interest containing the subcellular objects of interest.
NOTE: This protocol uses segments of a FIB-SEM image volume from late adolescent ferret primary visual cortex (24.2 x 16.2 x 2.4 µm, 4 nm isotropic voxels), and a freely accessible FIB-SEM volume from adult rat CA1 hippocampus (10.2 x 7.7 x 5.3 µm; 5 nm isotropic voxels) made available by the Knott laboratory (https://www.epfl.ch/labs/cvlab/data/data-em/). It is critical to prepare tissue blocks for FIB-SEM using a protocol that preserves ultrastructural detail and produces high contrast under FIB-SEM imaging conditions23,24. Importantly, the tissue should be imaged such that the resulting FIB-SEM image voxel resolution is <0.5 × the size of the diameter of the smallest subcellular structure of interest. For example, because the sites of spinule protrusion into presynaptic boutons can be as small as 30 nm in diameter, a voxel size of <15 nm would be appropriate to sample these structures above the Nyquist criterion. In addition, it is useful to image tissue at isotropic (equal in all dimensions) resolution (e.g., 5 x 5 x 5 nm voxels) to enable visualization and reconstruction of fine structures that have equal dimensions in all x/y/z planes. - Download and install the Fiji distribution of ImageJ25 (https://imagej.net/software/fiji/downloads).
- Align the image volume.
NOTE: While modern FIB-SEM systems produce roughly aligned image volumes, image to image registration may still suffer from sub-pixel drift.- In Fiji, navigate to Plugins | Registration and choose Register Virtual Stack Slices. Consult the User Manual for assistance (https://imagej.net/Register_Virtual_Stack_Slices). Start with a Rigid registration and then use an Affine technique if required.
- Scale the image volume.
- Open the aligned image volume in Fiji by clicking on File | Import | Image Sequence and navigating to the folder with the aligned images.
- In the dialog window, click on Sort names numerically if the images are named in numerical order. Click on Use virtual stack for faster Fiji performance.
- Once the image is open, click on Image | Properties and insert the physical size of the image volume pixels in micrometers in the pixel height, width, and depth boxes. Type um OR µm in each of the rightmost boxes. Save this aligned image volume as a .tiff file by clicking File | Save As | Tiff.
NOTE: If the image was scaled, the top of the image should now indicate the size of the image in micrometers (e.g., 9.82 x 8.65 µm).
2. Neurite and synaptic spinule identification
- While scrolling through the stack (using the arrow keys or mouse wheel), refer to the neurite identification decision tree (Figure 1) and examples of sectioned neurites (Figure 2) to locate and identify the objects of interest.
- Identify synaptic spinules as membrane-bound protrusions from a neurite that becomes fully enveloped within a presynaptic or postsynaptic neurite.
NOTE: Positive synaptic spinule identification requires visual evidence of the spinule-projecting object membrane invaginating into the inverted membrane of a presynaptic or postsynaptic structure and the presence of the membrane-bound spinule fully encapsulated by the pre- or postsynaptic structure's membrane in at least one image.
- Identify synaptic spinules as membrane-bound protrusions from a neurite that becomes fully enveloped within a presynaptic or postsynaptic neurite.
- Making regions of interest (ROIs) in Fiji
- Click on an appropriate tool in the Fiji toolbar (e.g., oval tool) and then click and drag on the image to form an ROI. Record this ROI in the ROI Manager by typing the letter "T."
NOTE: The ROI Manager should now appear with the image coordinates as its default name. - Rename the ROI as desired by clicking the Rename button in the ROI Manager and typing the desired name in the text box that appears. Rename ROIs based on quantitative criteria (e.g., the 12th presynaptic bouton that is analyzed and has two synapses with postsynaptic spines might be named B12_s,s).
NOTE: To associate each ROI with one image "slice" (rather than have it present in the entire image stack) and to make ROI names visible on single images, click on these options in the ROI Manager | More | Options window and then click the Show All and Labels check boxes at the bottom of the ROI Manager. Note that this step is crucial when saving ROIs within an image volume because associating ROIs with the entire image volume rather than individual images can lead to double-counting of objects within the volume. - To save all the ROIs currently in the ROI manager as a file, first, click the Deselect button in the ROI Manager, click the More button, and scroll down to Save.
NOTE: This saved .zip file can now be opened in Fiji to reload all of these ROIs into the ROI Manager.
- Click on an appropriate tool in the Fiji toolbar (e.g., oval tool) and then click and drag on the image to form an ROI. Record this ROI in the ROI Manager by typing the letter "T."
3. Determine the area of interest and transfer image volume to 3D analysis software
- Determine whether it is desirable to transfer the entire image volume to the 3D reconstruction and analysis software, or if a smaller substack (see 3.3.2) should be created to analyze a smaller number of images and/or region of interest from the original stack.
- Run a small 'pilot' study on a relatively small area containing the object(s) of interest to estimate effect size and the number of objects of interest required to obtain the desired power for statistical comparisons. Use a conservative estimate of the size of the image volume required to sample the objects of interest if these subcellular objects are non-uniformly distributed within the image volume.
- To use a portion of the image volume for 3D analysis, first duplicate the image volume by clicking on Image | Duplicate.
- To use a cropped portion of the image for analysis, choose the rectangle tool from the Fiji tool bar and draw the desired region of interest; if this is not desired, continue to the next step.
- Go to Image | Stacks | Tools | Make Substack. Type in the range of images to include within the substack (e.g., include images 1 to 500 by typing 1 – 500) in the text box.
- Refer to the sampling considerations above (note after 1.1) to determine the required z-resolution based on the size of the object(s) of interest. For example, if the image volume has a voxel depth of 5 nm, it is possible to sample the object of interest at 10 nm intervals in the z-dimension so that Fiji retains every other image in the substack. To do this, type the number of the nth image after the range (e.g., to only include every other image in a 1 – 500 image range, type 1 – 500 – 2).
NOTE: Each 3D analysis software package (see Table 1) will have its own file import requirements. This protocol focuses on importing image volumes saved as .tiff files into the image volume software, Reconstruct.
4. 3D reconstructions and analysis of thin subcellular structures in Reconstruct
NOTE: It is highly advantageous to use a mouse equipped with a wheel while using Reconstruct. In addition, if most or all traces will be performed manually, using a stylus to draw outlines on a computer with a touchscreen can dramatically increase trace efficiency.
- Download and install Reconstruct (https://synapseweb.clm.utexas.edu/software-0) for use on a computer running Microsoft Windows in a native or virtual environment.
- Start a new reconstruction by creating a new folder for the reconstruction files, clicking on Series | New, and saving this reconstruction (.ser file) with the desired file name in the folder that was just created.
- Import the saved .tiff image volume into Reconstruct.
- Navigate to Series | Import | Images, click Select in the Import Images window, and locate the image volume file. In the Import Images window, look for the image volume files that appear as a separate line for each image in the volume.
- Important: Type the correct pixel size in the Pixel Size text box to correspond to the x/y pixel size (in µm) of the image volume. Once this is done, click on the Import button in the Import Images window.
NOTE: Once the images have been imported, there should be an image in the middle of the Reconstruct window, and the Import Images list should be empty. - If there is an image in the middle of the Reconstruct window and the Import Images list is empty, click on Quit in the Import Images window.
- Set the section thickness (i.e., voxel height or z-dimension) by clicking on Section | List sections, highlighting all of the listed sections, then clicking on Modify | Thickness in the Section List menu and typing the section thickness (in µm) into the text box (e.g., 0.005 for a 5 nm thick section).
NOTE: Recall that if working with a substack from a larger original image volume that only includes every nth section (see step 3.3.3), it is necessary to account for these 'missing' sections in the total section thickness. For example, if the original image volume section thickness (aka voxel height) was 0.005 µm, and the substack only includes every third section (e.g., 1-500-3), the section thickness of this substack is 0.015 µm (i.e., 0.005 µm x 3 sections).
- Make traces of the objects of interest.
- Go to Series | Options | Names/Colors Tab to give the new object a name. Pick a color for the outline and interior fill of the object's traces, and then click OK.
NOTE: Reconstruct will attempt to connect all traces with the same name as one object. Refer to the Reconstruct manual for trace naming conventions (e.g., do not include spaces in names). Trace colors are merely for visualizing traces; the color of this object's 3D reconstruction can be easily altered later. - On the Toolbar, select the Draw Freehand tool that looks like a pencil. Hold down the left mouse button and drag the mouse to trace the object's membrane; let go of the left mouse button to autocomplete the trace. Click Page Up / Page Down or use the mouse wheel to advance to the next optical section in the image volume and progressively make traces until the structure is complete.
- Go to Series | Options | Names/Colors Tab to give the new object a name. Pick a color for the outline and interior fill of the object's traces, and then click OK.
- Tracing synaptic spinules
NOTE: Embedded spinules within presynaptic boutons appear as membrane-bound transparent structures, occasionally with actin-like electron-opaque material and/or membrane-like folds in their interior.- Advance through the image volume and locate the site of spinule invagination into the presynaptic bouton, being careful to differentiate spinules from endoplasmic reticula, vacuoles, endosomes, autophagosomes, and other subcellular organelles.
NOTE: Unlike membrane-bound organelles, ensure that synaptic spinules always project from (i.e., everted membrane) a neurite/glia and are received by (i.e., inverted membrane) a pre- or postsynaptic structure. This visual evidence of spinule invagination should occur in ≥2 images, and a fully encapsulated membrane-bound spinule should be present within the pre- or postsynaptic structure in ≥1 image. Small spinules may be clathrin-coated. To differentiate spinule invagination events from clathrin-coated endocytosis into the pre- or postsynaptic neurite, ensure that positive identification of clathrin-coated spinules requires visual confirmation of the coincident everting membrane from the spinule parent object projecting into the inverting membrane of a pre- or postsynaptic structure within the same image. - Advance through the image volume to locate the site of spinule projection/origin from its parent neurite/glia; refer to Figure 1 and Figure 2 to help identify these parent structures.
- Begin the trace of the spinule at the site of membrane protrusion into the receiving structure (e.g., presynaptic bouton) and continue drawing the spinule for as long as it appears as a membrane-bound structure within the encapsulating pre- or postsynaptic neurite.
- Continue holding down the left mouse button (or keep drawing along the spinule membrane with a stylus if using a touch screen computer) while tracing the membrane of the spinule with the mouse pointer.
- When the point where the trace started is reached, let go of the left mouse button.
NOTE: Letting go of the left mouse button too soon will autocomplete the trace, often with undesirable results. If this happens, simply hit delete while this trace is highlighted in the Trace List. - Repeat steps 4.5.2-4.5.5 for every section where the spinule appears.
NOTE: Remember to trace the spinule as an encapsulated membrane-bound structure within the bouton when appropriate. Because Reconstruct does not permit the automated subtraction of encapsulated subcellular structures (e.g., spinules, subcellular organelles) from the encapsulating structure's volume, this can be performed post-hoc (e.g., by simply subtracting the spinule volume from the presynaptic bouton's total volume).
- Advance through the image volume and locate the site of spinule invagination into the presynaptic bouton, being careful to differentiate spinules from endoplasmic reticula, vacuoles, endosomes, autophagosomes, and other subcellular organelles.
- Reconstructing objects in 3D in Reconstruct
- Navigate to Series | Options | 3D tab, and ensure the Traces radio button is clicked.
- Open the Object List by clicking Objects | List objects.
- Once the names of the traced object(s) load in the window, double-click on the name of the object(s) in the Object List to view them in 3D. Wait for the 3D Scene window to appear.
- Ensure that the start, end, and counts of the traces are correct in the Object List for each object, such that the Count = End – Start. If the Count ≠ End – Start for an object, then one or more traces from this object may have mistakenly been omitted.
- Rotate the 3D traces of the object(s) by holding down the left mouse button and moving the mouse and/or holding down the right mouse button and moving the mouse forward and backward to zoom in and out. Ensure that the object's traces are appropriately aligned and that there appears to be a smooth transition between each successive trace.
- If necessary, retrace individual traces to remove errors that lead to sharp edges and/or transitions between traces.
- Put a tetrahedral mesh skin on the object by navigating to Series | Object | 3D tab and clicking on the Boissonnat surface radio button.
- In the 3D Scene window, click on Scene | Clear, then double-click on the desired object name in the Object List.
- Rotate the object(s) to ensure smooth transitions between traces and redraw traces if necessary.
- To change object colors and transparency, click on the name of the object in the Object List, and then click on Scene | Color/transparency… in the Object List menu. Double-click on the object name in Object List to view this change in the 3D Scene window.
NOTE: Object color, transparency, and smoothing can be altered with a greater degree of control and specificity using Blender (see 5.4-5.6).
- Quantitative analysis of 3D objects
- Navigate to Series | Options | Lists tab and click on all the desired measurements in the Object List and Trace List sections of this window.
- Export the object measurements by opening the Object List (Object | List objects…) and then clicking on List | Save.
- Save the object measurements as displayed in the Object List as a comma delimited file (.csv).
- Import the .csv file into the data analysis software of choice. To import into a spreadsheet (e.g., Microsoft Excel), simply double-click the file, or open a new blank workbook, click on File | Import | CSV file option, and follow the instructions.
- To measure the distances between objects in 3D (e.g., to measure the projection distance of a synaptic spinule within a presynaptic bouton), use the Z-trace function in Reconstruct.
- First, give the trace a new name by navigating to the Series | Options | Names/Colors tab and typing in a name for the Z-trace.
- Next, click on the Z-trace tool in the toolbar.
- To draw the Z-trace of the projection distance of a synaptic spinule, click on the start of the synaptic spinule by clicking on the left mouse button, and then press the Page Up or Page Down buttons to advance through the image volume while clicking on successive points along the synaptic spinule's membrane within each section.
- Complete this Z-trace by clicking on the right mouse button.
- Export Z-trace measurements by clicking on Trace | List traces… to bring up the Trace List and then clicking on List | Save to save the trace list measurements as a .csv file.
- Adding a scale cube
- Navigate to one of the first or last images in the stack
- Go to Series | Options | Names/Colors tab and type a name for the scale cube.
- In the toolbar, pick the Draw Rectangle tool and draw a small square (size unimportant) adjacent to the objects but not obscuring them.Go to Series | Options | 3D tab, click on the Box radio button option in the Generate area, and specify the size of the scale cube (in µm) by typing its dimensions in the A:, B:, C: boxes in the Size area (e.g., type 0.5 for A, B, and C to generate a 0.5 µm/side scale cube).
NOTE: Before attempting to reconstruct the scale cube in 3D, be sure to have the desired 3D objects already reconstructed in the 3D Scene using the Traces or Boissonnat surface option. Remember to change the Generate option back to Boissonnat surface and the sizes of A, B, and C back to -1 after reconstructing the scale cube (i.e., after step 4.8.5). - Open the Object List and double-click on the name of the scale cube; wait for the scale cube to appear in the 3D scene window.
- To move the scale cube relative to the other objects, go to the Series | Options | 3D tab and type the number of µm desired to move the scale cube into the X, Y, and/or Z boxes. Click OK to close the window, and then double-click on the nom of the scale cube in the Object List.
NOTE: Alternatively, this can be done using Blender (see below).
5. Importing, recoloring, smoothing, and adding transparency to 3D reconstructions in Blender
NOTE: For detailed assistance in using Blender, please consult the Blender manual and/or the myriad "how to" videos on using each Blender function (simply do a web search for Blender AND Desired Function). What follows is a short primer on how to recolor, smooth, and add transparency to 3D reconstructions.
- Download and install the latest version of Blender, the free, open-source 3D software suite (www.blender.org).
- In Reconstruct, save the 3D reconstructions by bringing up the desired object(s) in the 3D scene window and then clicking Scene | Export As | VRML 2.0.
- In Blender, navigate to File | Import | X3D Extensible 3D (.x3d/.wrl). Click on the VRML 2.0 file that was saved from Reconstruct and then click on the Import X3D/VRML2 button.
- Recolor the reconstructions.
- In the Scene Collection window, in the upper right corner, click to deselect/uncheck the eye icon for the default Cube object, and click on the names of the individual objects to highlight each object in the scene. Rename these objects as appropriate.
- Click on the name of the first object to recolor in the Scene Collection window.
- Click on the circular Context. Material icon on the bottom of the righthand strip of icons, and then click Add Material. A list of options should appear below. Next, click on the bar next to the Base Color option and choose the desired luminance and hue for the object.
- To alter the metallic appearance, shininess, and reflective qualities of the object, adjust the Metallic, Specular, Roughness, and Sheen options to produce the desired effect.
NOTE: For certain options to appear in the Scene (e.g., transparency, sheen), it may be necessary to be in the Viewport Shading: Look Dev or Viewport Shading: Rendered viewport setting. To change to either viewport setting, hover over the globe icons in the upper right and click on the desired viewport setting. - To change the lighting angle, click on the Light object in the Scene Collection window, click on the Show Gizmo icon on the top right, and then drag the light around the scene to achieve the desired lighting effect. Change the light intensity in the Object Data window that appears underneath the Scene Collection window when the Light object is selected.
- To rotate objects in select axes separately, click on the names of the objects in the Scene Collection window, hit the R key, then click on the letter of the axis of rotation (x, y, or z key), and move the mouse to produce the desired rotation. To grab and move objects, select them in the Scene Collection window, hit the G key, and then use the mouse to move objects around the scene.
- Remesh (i.e., smooth) the reconstructions.
- Click on the name of the object of interest from the Scene Collection window and then use the gimball and mouse wheel to center and zoom in on the object.
- Click on the Context.Modifiers wrench icon on the righthand strip of icons, click Add Modifier, and then Remesh (under the Generate heading). Use the dropdown menu to select Smooth, click on the Smooth Shading button, and change the Octree Depth and Scale to achieve the desired smoothness.
NOTE: It is helpful to be in the Viewport Shading: Wireframe window (globe icon option in upper right) while remeshing to view the results directly. Remeshing will create different-sized tetrahedral meshes on the object. While smoothing the object to remove sectioning or tracing artifacts, be careful to retain its original shape and contours. Start at an Octree Depth = 7 and a Scale = 0.95, and then decrease these numbers slowly if necessary. Pay particular attention to small, thin objects, such as spinules and postsynaptic densities, as the small volume of these objects decreases the range of smoothing that can be done without altering their original shape.
- Add transparency to objects.
NOTE: Making objects transparent is important when it is desirable to show the 3D relationship of one object (e.g., spinules, presynaptic vesicles, mitochondria, endosomes) residing within another object (e.g., a presynaptic bouton).- Click on the desired object name in the Scene Collection window. Scroll to the bottom of the Context. Material window (bottom right icon in righthand list), open the Settings section, and use the dropdown menu to change the Blend Mode à Alpha Blend. Next, open the Surface section, scroll up to Alpha, and change the number to a value less than one (1 = fully opaque, 0 = fully transparent).
- Export snapshots of the 3D reconstructions.
- Once the optimal coloring, smoothing, and lighting for the 3D reconstructions have been obtained, save the image within the scene by clicking on the menu heading Window, and selecting Save Screenshot.
- In the Save Screenshot options on the lower left, choose the file type, color format, and compression desired.
- Type the nom for the new screenshot in the file name text box above, and then save the screenshot in the desired location.
Representative Results
Quantifying the percentage of synaptic spinules within the excitatory presynaptic bouton population in ferret primary visual cortex
Although spinule-like protrusions from neurites into excitatory presynaptic boutons have been observed for decades19,26, their potential importance for synaptic function has remained obscure. These experiments were designed to determine the proportion of excitatory presynaptic boutons containing spinules throughout postnatal development in the ferret primary visual cortex (V1) to ascertain the potential importance of spinules for synaptic function in relation to developmental milestones. Accordingly, aligned 4 nm/voxel isotropic FIB-SEM image volumes were acquired from a postnatal day (p)21 (15.1 x 14.1x 2.8 µm), p46 (9.7 x 8.4 x 2.7 µm), p60 (24.2 x 16.2 x 2.4 µm), and >p90 (24.2 x 16.2 x 2.4 µm) ferret V1, imaged using an FEI Helios 660 DualBeam FIB-SEM at 52° tilt, 4.2 mm working distance, 3 kV acceleration voltage, and 400 pA current in backscatter mode. Although stacks were roughly aligned with FEI software on acquisition, all stacks went through subpixel alignment using Fiji (refer to 1.4.3). These ages correspond to before the onset of correlated visual experience (i.e., eye-opening; p21), at the height of the canonical critical period for ocular dominance plasticity in ferret V1 (p46), near the end of the critical period (p60), and late adolescence (>p90).
Each image volume was scaled using Fiji, and every excitatory presynaptic bouton within the four FIB-SEM volumes was identified. Excitatory synapses were identified by the presence of parallel presynaptic and postsynaptic membranes, a prominent asymmetric (Gray's Type I) postsynaptic density27, and ≥3 presynaptic vesicles. Every excitatory presynaptic bouton was evaluated across its 3D volume for the presence of one or more spinules based on conservative criteria (refer to 4.5) and to determine whether the postsynaptic density (PSD) had a discontinuity termed a perforation that is associated with higher rates of plasticity28,29. In addition, each spinule was followed back to its parent structure to determine the proportion of spinules that emanate from distinct neurites or glia. Toward this end, regions of interest (ROIs) were outlined using the oval tool in Fiji for each excitatory presynaptic bouton by encircling the entire synapse (see 2.2.1 – 2.2.3). Each ROI contained the number of the boutons (in sequential order) and noted the presence/absence of a spinule, the postsynaptic target (e.g., dendritic spine or dendritic shaft), and the presence/absence of a perforated PSD (see Note after 2.2.3). By examining every excitatory bouton within these four image volumes, the percentages of presynaptic boutons that contained spinules were determined to increase across development. Yet, the 3D relationships of spinules to presynaptic boutons remained to be determined.
Examining the relationship between synaptic spinules and their encapsulating excitatory presynaptic boutons
To examine the relationship between the two most abundant spinule types (i.e., spinules emanating from postsynaptic spines and those projecting from adjacent axons) and their encapsulating excitatory spinule-bearing presynaptic boutons (SBBs), the >p90 image volume was analyzed to determine the sizes of these spinule types and whether these spinules were engulfed by similar-sized SBBs. Using the ROIs from the spinule prevalence quantification in Fiji described above, SBBs that contained spinules from postsynaptic spines or adjacent axons/boutons were identified. After examining these SBBs and spinules in 3D within Fiji, it was determined that a section thickness of 8 nm was sufficient to resolve the thin invagination of each spinule into its respective SBB. As such, substacks containing every other section (i.e., 8 nm z resolution) were made, with a substack range that included the full 3D extent of SBBs and their spinules (see 3.3). After transferring these substacks to Reconstruct, 11 SBBs containing postsynaptic spine spinules, and 14 SBBs containing adjacent axons spinules were traced and three-dimensionally reconstructed (see 4.4-4.7). These analyses revealed that postsynaptic spine spinules were 2.7 times larger than adjacent axon spinules (0.016 ± 0.005 µm3 vs. 0.0059 ± 0.001 µm3, mean ± SEM, postsynaptic spine vs. adjacent axon spinules, respectively; Figure 3C). However, given the small sample sizes for this pilot study, these data were not statistically significant at p < 0.05 (Mann Whitney U test, two-tailed, U = 56, p = 0.26). Using a freely available effect size and statistical power analysis software (G*Power)30, it is estimated that if this medium effect size (0.596) for the difference in spinule volumes holds, ~60 more postsynaptic spine and adjacent axon spinules reconstructions will be needed to obtain a statistically significant result with α = 0.05 and statistical power of 0.95. Interestingly, these analyses also found that while the volumes of SBBs containing postsynaptic spine spinules were similar to those of SBBs containing adjacent axon spinules (0.21 ± 0.04 µm3 vs. 0.18 ± 0.02 µm3, postsynaptic spine vs. adjacent axon containing SBBs, respectively), the volume that adjacent axon spinules occupied within their enveloping SBBs was nearly identical to the volume that postsynaptic spine spinules occupied within their SBBs (19.3 ± 3.2 % vs. 17.5 ± 2.4 %, postsynaptic spine vs. adjacent axon spinules, respectively; Mann Whitney U test, two-tailed, U = 68, p = 0.64; Figure 3D). In sum, these pilot data suggest that postsynaptic spine spinules may be larger than their adjacent axon counterparts, and that adjacent axon spinules may preferentially invaginate into a population of relatively small boutons in ferret V1.
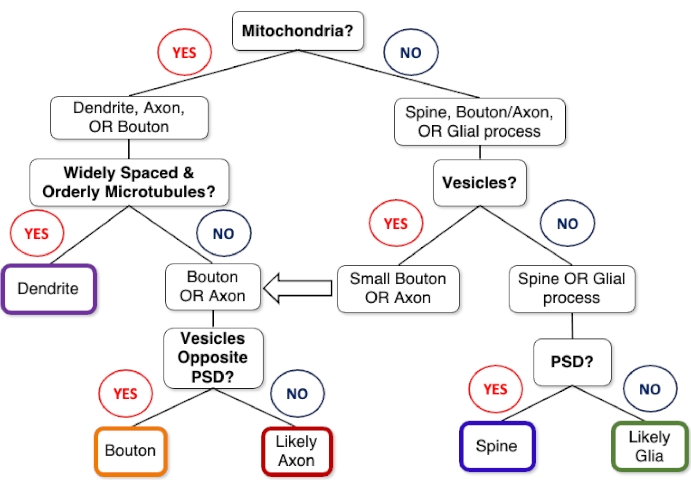
Figure 1: Decision tree for identifying neurites within FIB-SEM images. Neurite cross-sections (e.g., longitudinal and transverse sections) within FIB-SEM images can be identified based on the presence/absence of a few key organelles. For example, neurites containing mitochondria include dendrites, axons, and presynaptic boutons. Yet, under most FIB-SEM staining protocols, only dendrites will have a prominent, widely spaced (~55-70 nm spacing)31, and orderly arrangement of microtubules. In contrast, axons and boutons display neurotransmitter-containing vesicles, with boutons exhibiting a dense pool of these synaptic vesicles at their active zone(s) opposite the PSD, while axons contain dense (~13-30 nm spaced)31 microtubules and lower contrast neurofilaments. Dendritic spines nearly always lack mitochondria, microtubules, and vesicles, and therefore can most readily be differentiated from glial processes (e.g., astrocytes) based on the presence of a PSD. However, spines are also mostly larger than glial processes, connect to their parent dendrite, and sometimes contain a spine apparatus. Abbreviations: FIB-SEM = focused ion beam scanning electron microscopy; PSD = postsynaptic density. Please click here to view a larger version of this figure.
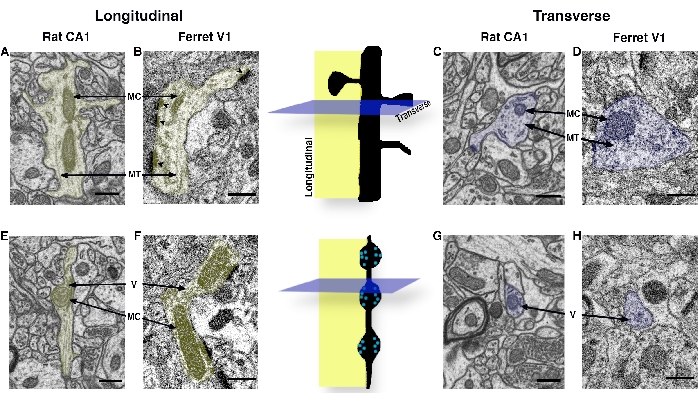
Figure 2: Neurite cross-section identification primer, displaying neuronal dendrites and axons from adult rat CA1 hippocampus and late adolescent ferret V1. As neuronal dendrites and axons have a range of sizes and course through the brain at angles tangential to the plane of sectioning, similar-sized dendrites and axons cut at different angles have unique appearances within FIB-SEM images. Central cartoons show a dendrite (above) and an axon (below) sectioned in a longitudinal plane (yellow) and a transverse plane (blue). (A, B) Neuronal dendrites sectioned in a longitudinal plane (i.e., along their long axis). Arrows point to distinguishing mitochondria (MC) and orderly widely spaced 'stripe-like' microtubules (MT). Note that dendrites followed along their depth should also exhibit PSD (arrowheads; dark, electron-dense regions along dendrite and spine in B). (C, D) Neuronal dendrites sectioned in a transverse plane (i.e., along their short axis). Arrows point to MT cut in a transverse plane that appear vesicular and MC that appear circular or ovoid. Dendrites can be differentiated from transversely-sectioned axons by their orderly, widely-spaced microtubules, ~1.25-2.75 × larger diameter32, absence of synaptic vesicles, and the presence of one or more PSDs. (E, F) Neuronal axons sectioned along a longitudinal plane, displaying MC and vesicles (V). Note that the axon in E contains orderly, densely-spaced microtubules, potentially leading to its misidentification as a dendrite, yet it also contains prominent neurotransmitter-containing vesicles. Clustered synaptic vesicles are also seen in the axon in F at two active zones opposite PSDs. Axons followed through the depth of most image volumes will display en passant synaptic boutons along their length, as in F. (G, H) Neuronal axons sectioned in a transverse plane, displaying prominent vesicles. Axons sectioned in a transverse plane often appear ovoid and have regions along their depth that are among the smallest diameter neuronal structures in the neuropil. Scale bars in A–H = 0.5 µm. Abbreviations: FIB-SEM = focused ion beam scanning electron microscopy; PSD = postsynaptic density; MC = mitochondria; MT = microtubules; V = vesicles. Please click here to view a larger version of this figure.
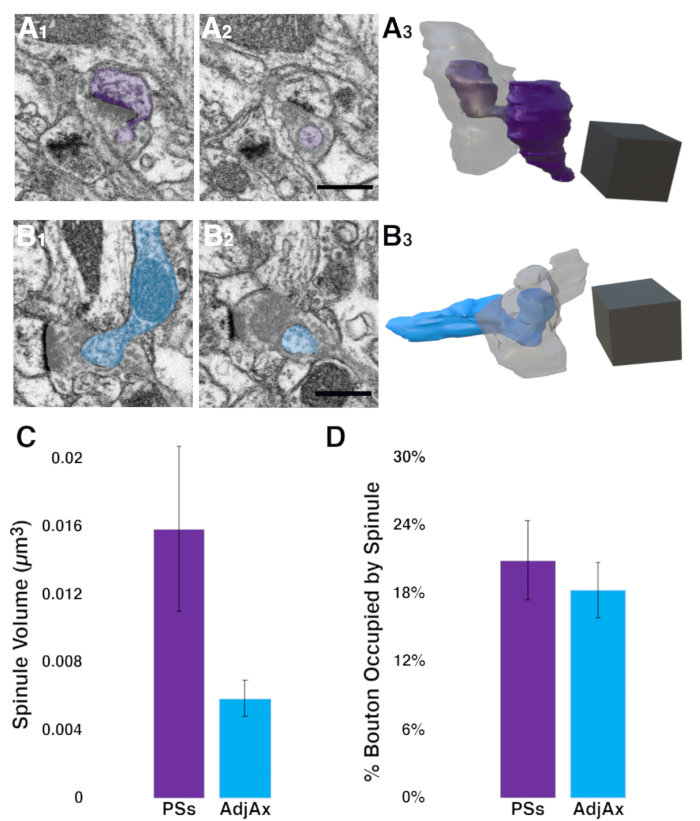
Figure 3. Pilot study using the described analysis pipeline to quantify postsynaptic spine and adjacent axon spinule volumes within presynaptic boutons in ferret V1. (A1–A3) Two sequential images showing the invagination (A1) and envelopment (A2) of a postsynaptic spine spinule (purple) into an excitatory presynaptic bouton (gray). Full 3D reconstruction (A3) showing the postsynaptic spine (purple) projecting a large anchor-like spinule into its presynaptic bouton (gray, made transparent) partner. (B1–B3) Two sequential images showing an adjacent (i.e., non-synaptic) axon (cyan) invaginating into (B1) and becoming fully encapsulated (B2) within a presynaptic bouton (gray). Note that the presynaptic bouton has a synapse with a postsynaptic spine to the left of both images, indicated by the prominent asymmetric PSD. Full 3D reconstruction (B3) of this adjacent axon spinule (cyan) within a presynaptic bouton (gray, made transparent). (C) Postsynaptic spine spinules show a trend toward being larger than spinules projecting from adjacent axons (n = 11 and 14, for PSs and AdjAx spinules, respectively; error bars indicate Standard Error of the Mean (SEM); Mann Whitney U test; p =0.26). (D) AdjAx spinules occupy a similar portion of their presynaptic boutons as PSs spinules (error bars indicate SEM; Mann Whitney U test, p = 0.64.). Scale bars for A2 and B2 = 0.5 µm; scale cubes for A3 and B3 = 0.5 µm/side. Abbreviations: PSD = postsynaptic density; PSs = postsynaptic spine spinules (PSs); AdjAx = adjacent axons. Please click here to view a larger version of this figure.
| Software | OS Compatibility | Semi-automated Segmentation | User-friendly UX | 3D Measurement Features | Download Site | |
| Espina | Windows; Linux | +++ | +++ | ++++ | https://cajalbbp.es/espina/#started | |
| IMOD | Windows; Linux; Mac | + | + | ++++ | https://bio3d.colorado.edu/imod/ | |
| Neuromorph (Blender plugin) | Windows; Linux; Mac | + | ++ | +++ | https://neuromorph.epfl.ch/software/ | |
| Reconstruct | Windows | + | ++++ | ++ | https://synapseweb.clm.utexas.edu/software-0 | |
| TrakEM2 (ImageJ Plugin) | Windows; Linux; Mac | ++ | +++ | +++ | https://www.ini.uzh.ch/~acardona/trakem2.html | |
Table 1: 3D Image volume analysis software. Selection of free, open-source software programs for registration, visualization, reconstruction, and measurement of thin subcellular structures within FIB-SEM image volumes. A qualitative assessment is presented for the degree to which each software platform contains three features salient for users (semi-automated segmentation, user-friendly user interface (UX), and 3D measurement features), with "++++" as the highest degree/amount, and "+" as the lowest degree/amount. Abbreviation: FIB-SEM = focused ion beam scanning electron microscopy.
Discussion
This FIB-SEM image volume analysis pipeline can produce reliable 3D reconstructions and quantitative measurements of thin subcellular structures. While current semi-automated techniques using deep neural network and segmentation algorithms can increase the speed and efficiency in reconstructing cellular structures possessing relatively high membrane contrast within large image volumes33, many subcellular structures (e.g., spinules, smooth endoplasmic reticula, endosomes) will remain difficult to reliably capture using automated methods due to their lower membrane contrast and/or tortuosity, though newer methods have begun to capture higher-contrast mitochondria and rough endoplasmic reticula34,35. Moreover, the subtle distinctions between unlabeled subcellular structures (e.g., clathrin-mediated endocytosis vs. spinule with clathrin-coated tip, or endosome vs. endoplasmic reticulum) will necessitate recurrent manual proofreading of any automated reconstruction technique36. Thus, this protocol details the importance of establishing clear criteria for the positive identification of subcellular objects prior to initiating an analysis of subcellular structures within FIB-SEM images.
Toward these ends, it is critical to become familiar with the appearance of a particular subcellular structure across a range of randomly sectioned angles (e.g., Figure 2) and imaging parameters to appropriately identify objects of interest and produce accurate and reliable traces of these structures within 3D image volumes. Moreover, the criteria and apparent structural diversity of a subcellular structure of interest must be agreed upon across all raters (i.e., analyzers) of the data through robust tests of interrater reliability using previously annotated ground truth data. For example, a suggested routine is to train raters on identical previously annotated image volumes until interrater reliability for subcellular object tracing is >95% before analyzing a new dataset.
It is also crucial to choose a FIB-SEM fixation and embedding protocol that maximizes the contrast of the object of interest. Each FIB-SEM freezing and/or fixation protocol has distinct advantages and challenges in its amenability to the imaging and localization of subcellular structures24,37,38. To facilitate spinule and other thin membrane-bound subcellular structure reconstructions from FIB-SEM image volumes, it is beneficial to use a fixation protocol that uses both formaldehyde and glutaraldehyde, long heavy metal and/or uranyl acetate incubations, and an epoxy resin embedding protocol4,37. While high-pressure freezing is the 'gold standard' technique for preserving native protein structure within thick tissue samples without inducing dehydration or protein cross-linking artifacts24,39, identifying and reconstructing thin membrane-bound structures within FIB-SEM images requires a protocol that delivers higher contrast than most freezing protocols afford.
This protocol focuses on identifying, reconstructing, and analyzing thin spinules that invaginate into, and become wholly encapsulated by, presynaptic boutons. Identifying and analyzing thin subcellular structures connected to a parent structure at one end, and embedded within a separate structure at its other end, offers unique challenges. For example, it is crucial to establish criteria for what constitutes an envelopment of the thin subcellular projection. While it is conceivable that one could establish envelopment as being nearly fully surrounded by another structure, as a peninsula in an ocean, this protocol requires that spinules are observed as fully embedded membrane-bound objects within enveloping presynaptic boutons (see 4.5 and Figure 3). This distinction separates thin structures that partially project into other objects from thin structures that invaginate and are surrounded by the enveloping object's membrane, as a hand clasped around a finger. Functionally, the latter case involves greater surface area contact, increased potential for projection-to-enveloping structure's organelles to interact, and a greater structural/energy investment between the thin invaginating subcellular projection and the enveloping object. In addition, when reconstructing and analyzing these thin embedded objects, it is important to determine whether the 3D reconstruction software can automatically subtract the volume of the thin embedded structures from the encompassing objects. For example, using Reconstruct, one can simply subtract the spinule volume from the bouton's volume post-hoc, producing a ~100% increase (after vs. before subtraction) in the spinule volume to bouton volume ratio.
Importantly, this protocol is focused on accurately and efficiently analyzing thin subcellular structures imaged with a small voxel size (e.g., <15 nm/voxel) that enables the faithful sampling of their 3D ultrastructure. As such, these steps are amenable to relatively rapid analysis of hundreds of subcellular structures within FIB-SEM image volumes within a reasonable time frame (e.g., weeks to months). However, as this protocol uses manual segmentation to three-dimensionally reconstruct objects of interest, it would be difficult and likely unreasonably time-consuming to identify and reconstruct thousands of thin subcellular objects for a single experiment. Moreover, if the subcellular objects of interest, or some subset of them, are relatively sparse within the image volume, identifying enough of these objects for statistical comparisons may be difficult. In these cases, it would be advantageous to write/use an algorithm to speed the identification and/or segmentation of the subcellular structures, followed by manual proofreading of the identification and segmentation process.
Quantitative analysis of thin subcellular structures within FIB-SEM image volumes is a robust, reliable, and adaptable technique, being used to investigate such disparate structures as the complex arrangement of an embedded Plasmodium chabaudi parasite within an erythrocyte40, the 3D organization of thin telopode projections within cardiac telocytes41, and the thin cytoplasmic processes of osteocytes and osteoblasts42. Here, an analysis pipeline is described that can serve as an entry point into the identification, reconstruction, and analysis of thin subcellular structures within FIB-SEM image volumes. While this protocol focused on the analysis of synaptic spinules within presynaptic boutons in brain tissue, these generalizable steps are amenable to the reconstruction and analysis of thin subcellular projections across a range of tissue and cell types.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by the University of Washington Bridge Fund and the University of Washington Tacoma Pilot RRF Fund. Many thanks to Dr. Claudia Lopez and Dr. Jessica Riesterer from the MMC at Oregon Health & Sciences University for FIB-SEM technical support, Dr. Graham Knott for the use of the CA1 FIB-SEM image volume, and the UW Tacoma students in the Neuronal Reconstructions (TBIOMD 495) course for their patience and excellence in working with this protocol.
Materials
| Fiji (ImageJ) | https://imagej.net/software/fiji/downloads | ||
| Reconstruct | https://synapseweb.clm.utexas.edu/software-0 | ||
| Blender | Blender Foundation | https:/www.blender.org |
References
- Holler, S., Kostinger, G., Martin, K. A. C., Schuhknecht, G. F. P., Stratford, K. J. Structure and function of a neocortical synapse. Nature. 591 (7848), 111-116 (2021).
- Xu, C. S., Pang, S., Hayworth, K. J., Hess, H. F., Wacker, I., Hummel, E., Burgold, S., Schroder, R. Transforming FIB-SEM. Volume microscopy: Multiscale imaging with photons, electrons, and ions. , 221-243 (2020).
- Merchan-Perez, A., Rodriguez, J. R., Alonso-Nanclares, L., Schertel, A., Defelipe, J. Counting synapses using FIB/SEM microscopy: a true revolution for ultrastructural volume reconstruction. Frontiers in Neuroanatomy. 3, 18 (2009).
- Wu, Y., et al. Contacts between the endoplasmic reticulum and other membranes in neurons. Proceedings of the National Academy of Sciences of the United States of America. 114 (24), 4859-4867 (2017).
- Weigel, A. V., et al. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell. 184 (9), 2412-2429 (2021).
- Rodriguez-Moreno, J., et al. Quantitative 3D ultrastructure of thalamocortical synapses from the “lemniscal” ventral posteromedial nucleus in mouse barrel cortex. Cerebral Cortex. 28 (9), 3159-3175 (2018).
- Khanmohammadi, M., Waagepetersen, R. P., Sporring, J. Analysis of shape and spatial interaction of synaptic vesicles using data from focused ion beam scanning electron microscopy (FIB-SEM). Frontiers in Neuroanatomy. 9, 116 (2015).
- Cali, C., et al. The effects of aging on neuropil structure in mouse somatosensory cortex-A 3D electron microscopy analysis of layer 1. PLoS One. 13 (7), 0198131 (2018).
- Vogelstein, J. T., et al. A community-developed open-source computational ecosystem for big neuro data. Nature Methods. 15 (11), 846-847 (2018).
- Morales, J., et al. Espina: A tool for the automated segmentation and counting of synapses in large stacks of electron microscopy images. Frontiers in Neuroanatomy. 5, 18 (2011).
- Kremer, J. R., Mastronarde, D. N., McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. Journal of Structural Biology. 116 (1), 71-76 (1996).
- Jorstad, A., Blanc, J., Knott, G. NeuroMorph: a software toolset for 3D analysis of neurite morphology and connectivity. Frontiers in Neuroanatomy. 12, 59 (2018).
- Fiala, J. C. Reconstruct: a free editor for serial section microscopy. Journal of Microscopy. 218, 52-61 (2005).
- Cardona, A., et al. TrakEM2 software for neural circuit reconstruction. PLoS One. 7 (6), 38011 (2012).
- Petralia, R. S., Wang, Y. X., Mattson, M. P., Yao, P. J. Invaginating structures in mammalian synapses. Frontiers in Synaptic Neuroscience. 10, 4 (2018).
- Petralia, R. S., Wang, Y. X., Mattson, M. P., Yao, P. J. Structure, distribution, and function of neuronal/synaptic spinules and related invaginating projections. Neuromolecular Medicine. 17 (3), 211-240 (2015).
- Pappas, G., Purpura, D. Fine structure of dendrites in the superficial neocortical neuropil. Experimental Neurology. 4, 507-530 (1961).
- Case, N. M., Gray, E. G., Young, J. Z. Ultrastructure and synaptic relations in the optic lobe of the brain of Eledone and Octopus. Journal of Ultrastructure Research. 39 (1), 115-123 (1972).
- Bailey, C. H., Thompson, E. B., Castellucci, V. F., Kandel, E. R. Ultrastructure of the synapses of sensory neurons that mediate the gill-withdrawal reflex in Aplysia. Journal of Neurocytology. 8 (4), 415-444 (1979).
- Wagner, H., Djamgoz, M. B. A. Spinules: a case for retinal synaptic plasticity. Trends in Neuroscience. 16 (6), 201-206 (1993).
- Campbell, C., Lindhartsen, S., Knyaz, A., Erisir, A., Nahmani, M. Cortical presynaptic boutons progressively engulf spinules as they mature. eNeuro. 7 (5), (2020).
- Spacek, J., Harris, K. M. Trans-endocytosis via spinules in adult rat hippocampus. Journal of Neuroscience. 24 (17), 4233-4241 (2004).
- Knott, G., Rosset, S., Cantoni, M. Focussed ion beam milling and scanning electron microscopy of brain tissue. Journal of Visualized Experiments: JoVE. (53), e2588 (2011).
- Steyer, A. M., Schertel, A., Nardis, C., Möbius, W. FIB-SEM of mouse nervous tissue: Fast and slow sample preparation. Methods in Cellular Biology. 152, 1-21 (2019).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Westfall, J. A. Ultrastructure of synapses in a primitive coelenterate. Journal of ultrastructure research. 32 (3), 237-246 (1970).
- Gray, E. G. Axo-somatic and axo-dendrritic synapses of the cerebral cortex: An electron microscope study. Journal of Anatomy. 93 (4), 420-433 (1959).
- Jones, D. G., Calverley, R. K. Perforated and non-perforated synapses in rat neocortex: three-dimensional reconstructions. Brain Research. 556, 247-258 (1991).
- Geinisman, Y., et al. Structural synaptic correlate of long-term potentiation: Formation of axospinous synapses with multiple, completely partitioned transmission zones. Hippocampus. 3 (4), 435-445 (1993).
- Faul, F., Erdfelder, E., Lang, A. -. G., Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behavior Research Methods. 39, 175-191 (2007).
- Chen, J., Kanai, Y., Cowan, N. J., Hirokawa, N. Projection domains of MAP2 and tau determine spacings between microtublules in dendrites and axons. Nature. 360, 674-677 (1992).
- Benavides-Piccione, R., et al. Differential structure of hippocampal CA1 pyramidal neurons in the human and mouse. Cerebral Cortex. 30 (2), 730-752 (2020).
- Kornfeld, J., Denk, W. Progress and remaining challenges in high-throughput volume electron microscopy. Current Opinion Neurobiology. 50, 261-267 (2018).
- Liu, J., et al. Automatic reconstruction of mitochondria and endoplasmic reticulum in electron microscopy volumes by deep learning. Frontiers in Neuroscience. 14, 599 (2020).
- Lucchi, A., Smith, K., Achanta, R., Knott, G., Fua, P. Supervoxel-based segmentation of mitochondria in EM image stacks with learned shape features. IEEE Transactions on Medical Imaging. 31 (2), 474-486 (2012).
- Chklovskii, D. B., Vitaladevuni, S., Scheffer, L. K. Semi-automated reconstruction of neural circuits using electron microscopy. Current Opinion in Neurobiology. 20 (5), 667-675 (2010).
- Knott, G., Marchman, H., Wall, D., Lich, B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. Journal of Neuroscience. 28 (12), 2959-2964 (2008).
- Takahashi-Nakazato, A., Parajuli, L. K., Iwasaki, H., Tanaka, S., Okabe, S., Burger, C., Velardo, M. J. Ultrastructural observation of glutamergic synapses by focused ion beam scanning electron microscopy (FIB-SEM). Glutamate Receptors: Methods and Protocols. , 17-27 (2019).
- Korogod, N., Petersen, C. C., Knott, G. W. Ultrastructural analysis of adult mouse neocortex comparing aldehyde perfusion with cryo fixation. Elife. 4, 05793 (2015).
- Soares Medeiros, L. C., De Souza, W., Jiao, C., Barrabin, H., Miranda, K. Visualizing the 3D architecture of multiple erythrocytes infected with Plasmodium at nanoscale by focused ion beam-scanning electron microscopy. PLoS One. 7 (3), 33445 (2012).
- Cretoiu, D., Hummel, E., Zimmermann, H., Gherghiceanu, M., Popescu, L. M. Human cardiac telocytes: 3D imaging by FIB-SEM tomography. Journal of Cellular and Molecular Medicine. 18 (11), 2157-2164 (2014).
- Hasegawa, T., et al. Three-dimensional ultrastructure of osteocytes assessed by focused ion beam-scanning electron microscopy (FIB-SEM). Histochemistry and Cell Biology. 149 (4), 423-432 (2018).

.
