N-glycan Profiling of Glycoproteins by Hydrophilic Interaction Liquid Chromatography with Fluorescence and Mass Spectrometric Detection
Summary
N-glycan profiling of glycoproteins is essential for discovering novel biomarkers and understanding glycan functions in cellular events. Additionally, N-glycan analysis of protein biopharmaceuticals is very important for human use. In this current article, a high-throughput strategy for identifying and quantifying N-glycan structures was presented using the HILIC-FLD-MS/MS technique.
Abstract
Glycosylation is a vital modification found in proteins. N-glycan profiling of glycoproteins is required to detect novel biomarker candidates and determine glycan alterations in diseases. Most commercially available biopharmaceutical proteins are glycoproteins. The efficacy of these drugs is affected by glycosylation patterns. Therefore, an in-depth characterization method for the N-glycans is necessary. Here, we present a comprehensive approach for qualitative and quantitative analysis of N-glycans using hydrophilic interaction liquid chromatography equipped with fluorescence detection and tandem mass spectrometry (HILIC-FLD-MS/MS). N-glycans were released from glycoproteins with a facile method and labeled by a procainamide fluorophore tag in the strategy. Subsequently, the procainamide labeled N-glycans were analyzed by a HILIC-FLD-MS/MS technique. In this approach, N-glycan structures were confirmed by the tandem mass spectrometric analysis, whereas fluorescence detection was used for the quantitative analysis. An application for data analysis of the detected N-glycan peaks is described in the study. This protocol can be applied to any glycoprotein extracted from various species.
Introduction
Glycosylation is a vital post-translational modification observed in proteins1. Multiple enzymatical processes regulate glycosylation modification in cellular organisms. Glycans are attached to the proteins by these enzymatical processes, and the proteins subjected to this modification are called glycoproteins1. Two glycosylation types are commonly observed in proteins. O-glycosylation is the attachment of O-glycans to the side chain of serine or threonine amino acid residues. N-glycosylation is the attachment of N-glycans to the side chain of asparagine amino acid residue in a protein.
The structure, stability, and folding of the proteins are affected by glycan attachments2. The glycosylation process dramatically influences the functions of the proteins, and glycoproteins regulate many cellular functions in organisms3,4. For example, heavily glycosylated proteins protect their glycoproteins from proteolytic degradation5. Another example is glycans of thyroid gland proteins that regulate Tg transport and hormone synthesis6,7. To explain their roles in cellular events, an in-depth characterization of glycoproteins is required8.
N-glycan profiles of the glycoproteins change in disease situations9,10,11,12. Profiling N-glycans derived from crucial glycoproteins or body fluids is required to discover novel biomarkers and understand the enzymatic activity changes in disease cases. On the other hand, most protein biopharmaceuticals are glycoproteins, and their glycan profiles influence drug efficacy13. Therefore, an acceptable method of N-glycan profiling must be performed in developing proper protein biopharmaceuticals for human use14.
Glycomics is an emerging discipline used to identify and quantify glycan structures of glycosylated molecules15,16. Many methods have been utilized for profiling the glycans of glycosylated species, including NMR17 and MS18. Hydrophilic Interaction Liquid Chromatography-with Fluorescence Detection (HPLC-HILIC-FLD) is the gold standard method for profiling N-glycans derived from glycoproteins19. When this strategy is combined with mass spectrometric detection, identifying N-glycan structures could be easier and more reliable. Most fluorescence tags used in N-glycan analysis with mass spectrometry have low ionization efficiencies. In contrast, procainamide increases the ionization efficiencies of N-glycans, which is used to obtain efficient tandem mass spectra of N-glycan structures20,21. Specific fragments can be obtained from this strategy by tandem mass spectrometry for the structural identification of N-glycans such as core fucosylated22 (proc-HexNAc1Fuc1) and bisecting types23 (proc-Hex1HexNAc3, proc-Hex1HexNAc3Fuc1).
This study demonstrates a facile protocol for the N-glycan profiling of glycoproteins with HILIC-FLD-MS/MS. The presented method includes four steps: (1) releasing of N-glycans from glycoproteins (2) labeling of N-glycans by a procainamide tag (3) purification of the procainamide labeled N-glycans, and (4) data analysis.
Protocol
NOTE: The human plasma used is commercially available (Table of Materials). No further biological samples obtained from humans were used.
1. Glycan release
- Denaturation of (glyco-)proteins
- Prepare the glycoprotein standards (e.g., IgG, a monoclonal antibody) at a concentration of a 10 µg·µL-1 in deionized H2O. For human plasma, the concentration used is 70 µg·µL-1.
NOTE: The samples should be vortexed until all solid proteins are dissolved. Human plasma (lyophilized) was used for the preparation of the plasma samples. - Take 20 µL of glycoprotein samples (200 µg) and lyophilized human plasma (1.4 mg).
- Add 40 µL of 2% SDS (sodium dodecyl sulphate) (w/v).
- Incubate the samples at 60 °C for 10 min to denature the (glyco-)proteins.
NOTE: The samples could be mixed at 500 rpm by a thermomixer during the incubation. A shaking water bath could be used alternatively.
- Prepare the glycoprotein standards (e.g., IgG, a monoclonal antibody) at a concentration of a 10 µg·µL-1 in deionized H2O. For human plasma, the concentration used is 70 µg·µL-1.
- Glycan release
- Add 20 µL of 4% Igepal-CA630 to the samples from step 1.1.4.
- Add 20 µL of 5x PBS (phosphate buffer saline).
- Prepare PNGase F enzyme at the concentration of 1 U·µL-1 in deionized water.
- Add 1 U of enzyme for glycoprotein standards and 2 U of enzyme for human plasma samples.
- Incubate the samples at 37 °C for 16 h.
- Procainamide labeling
- Prepare labeling solution using procainamide hydrochloric acid (110 mg·mL-1 in DMSO/AA (dimethyl sulfoxide /glacial acetic acid) 7/3, v/v).
- Prepare reductive amination solution using sodium cyanoborohydride (65 mg·mL-1 in DMSO/AA, 7/3, v/v).
CAUTION: Sodium cyanoborohydride is very toxic and flammable. Wear eye shields. 2-picoline borane complex (107 mg·mL-1 in DMSO) could be used alternatively. - Mix these solutions in a ratio of 1:1, v/v to prepare the labeling mixture.
- Add 100 µL of this labeling mixture to the glycan released sample from step 1.2.5.
- Incubate the samples at 65 °C for 2 h.
2. Purification of Procainamide Labeled N- glycans by Solid-phase Extraction (SPE) Cartridge
- Prepare a solution of microcrystalline cellulose (100 mg·mL-1) in deionized water.
- Take 300 µL of microcrystalline cellulose and insert it into the microcentrifuge tubes.
- Wash the microcrystalline cellulose with 1 mL of deionized H2O.
- Wash the microcrystalline cellulose with 1 mL of ACN/MQ (acetonitrile/dH2O), 85/15, v/v for conditioning.
NOTE: The microcentrifuge should be used for 1 min to discard washing solutions carefully. - Take 120 µL of glycan release solution and mix with 680 µL of ACN to obtain proper loading conditions (85/15, v/v, ACN/sample).
- Mix this mixture with microcrystalline cellulose in the microcentrifuge tubes.
- Incubate it at room temperature in a thermomixer by shaking at 500 rpm for 15 min.
- Transfer the slurry to an SPE cartridge (1 mL volume capacity).
NOTE: Do not allow air to enter cartridge packing. - Discard the loading solutions by passing slowly (1 drop/second).
NOTE: Ensure that the SPE system is connected to the vacuum pump. Solvents may flow with gravity. When necessary, apply vacuum via a vacuum pump. The applied vacuum pressure should be appropriately adjusted to transfer liquids down slowly. - Wash the sample by passing 1 mL of ACN/MQ/TFA (acetonitrile/dH2O/trifluoroacetic acid) solution (85/14/1, v/v/v) twice.
- Wash the sample by passing 1 mL of ACN/MQ mixture (85/15, v/v) twice.
- Elute the procainamide labeled N-glycans with 0.75 mL of water.
- Dry the elution solution with a concentrator overnight.
NOTE: Use a concentrator temperature of 45 °C for drying. - Dissolve the dried samples in a mixture of 100 µL of ACN/MQ (75/25, v/v) and transfer this solution to the vials, including an insert.
NOTE: The redissolved samples should be vortexed for 20 second.
3. HILIC-FLD-MS/MS Analysis
- Insert Tee (T) adaptors to the HILIC column to separate the flow into the two equal volumes. Connect one of both to the FLD detector, the other to the MS detector.
NOTE: The flow lines must be the same lengths. - Adjust a gradient program for HILIC separations of procainamide labeled N-glycans as indicated in Supplementary Figure 1. Use Mobile Phase A: 50 mM ammonium formate (pH: 4.4 and Mobile Phase B: 100 % ACN).
NOTE: The gradient program could be further optimized depending on the glycan heterogeneity of samples. - Click the Sampler button and set the injection volume to 10 µL.
- Click the Column Comp and adjust the column temperature to 60 °C.
- Click FLD and adjust the FLD excitation and emission wavelengths to 310 nm and 370 nm, respectively.
- Adjust the MS source, tune, and MS/MS parameters as displayed in Supplementary Figure 2.
NOTE: The MS and MS/MS parameters could be changed depending on the mass spectrometry used in the analysis.
4. Data Analysis
- Identification of procainamide labeled N-glycans
- Export MS/MS data by a proper format for database searches (e.g., .mgf, .xml).
- Insert them into a database search tool for the identification of N-glycans.
- Select the sample name listed in the software and click the Glycan Search button for the identification of procainamide labeled N-glycans by using the Carbbank database with given parameters (Supplementary Figure 3).
- Open the edit chromatograms button in the data analysis software for raw data and select extracted ion chromatogram type by filtering in all MSn.
- Add specific masses for core-fucosylated fragment proc-N1F1 (m/z 587.3+) and bisected fragments proc-H1N3 (m/z 1009.5+) and proc-H1N3F1 (m/z 1155.5+).
- Determine the precursors containing core-fucosylated and bisected N-glycan structure fragments.
- Export the identified N-glycan structures by the software (Supplementary Table 1-3).
- Check the precursors detected manually for determination of core-fucosylated and bisected N-glycan structures.
- Quantification of procainamide labeled N-glycans by an open-source software24
- Select FLD chromatogram in the chromatograms and click the method in the software. Open a method for exporting chromatograms as .xy file format.
- Convert the .xy file format of exported chromatograms to .txt files.
- Open the quantitative analysis software.
NOTE: The software used in the study can be downloaded from a Github source (https://github.com/Tarskin/HappyTools). - Follow the instructions and tutorials of the program presented by the developer.
NOTE: It is recommended to use a batch process to export relative areas. - Adjust the setting parameters (Supplementary Figure 4). Define four minimum peaks and 27 signal/noise ratios for calibrations.
- Open the results file with proper software such as Microsoft Excel for further evaluation.
Representative Results
In this presented approach, the N-glycans were first released, labeled by the procainamide tag and purified by cellulose-containing SPE cartridges. Then, N-glycan analysis of IgG, trastuzumab, and human plasma were performed by an HPLC-HILIC-FLD-MS/MS system. The MS (base peak) and FLD chromatograms of the determined N-glycan structures obtained from IgG and trastuzumab are shown in Figure 1, respectively. The MS/MS data obtained from these analyses were imported to the software and searched against a glycan database. Example MS/MS annotations of glycan structures from MS/MS spectra are given in Supplementary Figure 5. The lists of the detected N-glycan structures are provided in Supplementary Information Table S1 and S2 for IgG and trastuzumab, respectively. In addition, core fucosylated and bisected N-glycan types were detected by the analysis of fragment ions (proc-N1F1 (m/z 587.3+), proc-H1N3 (m/z 1009.5+) and proc-H1N3F1 (m/z 1155.5+) obtained from tandem MS analysis.
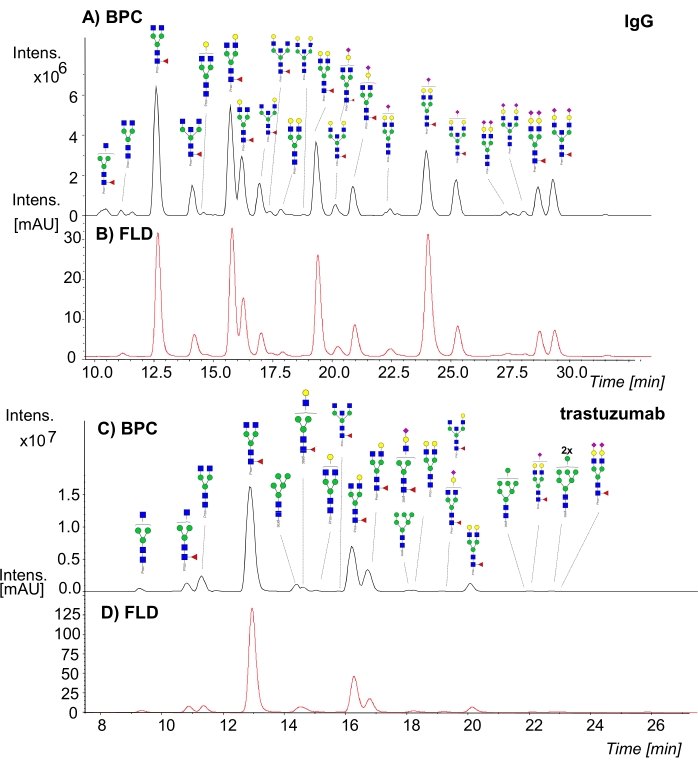
Figure 1: HILIC-FLD-MS/MS analysis of IgG and trastuzumab. Base peak and fluorescent chromatograms of procainamide-labeled N-glycans of (A, B) IgG and (C, D) trastuzumab, respectively. Please click here to view a larger version of this figure.
This strategy was also followed by the analysis of human plasma glycoproteome (Figure 2). The list of the N-glycans were listed in Supplementary Table S3 including the core fucosylation and bisecting N-glycan information. In the strategy, a python-based open-source tool was used to quantify N-glycan structures by using FLD chromatograms. The N-glycan profile of IgG N-glycans were exemplified and displayed in Figure 3. Thus, N-glycan profiling of glycoproteins were achieved by HILIC-FLD-MS/MS analysis.

Figure 2: HILIC-FLD-MS/MS analysis of human plasma glycome. (A) Base peak chromatogram (B) fluorescent chromatogram. Please click here to view a larger version of this figure.
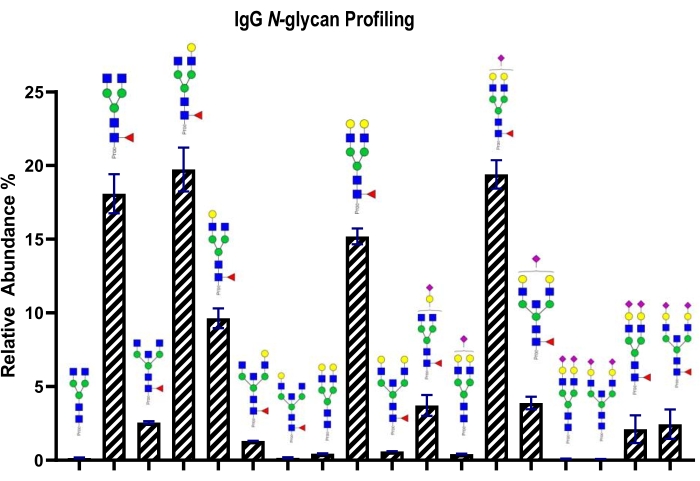
Figure 3: Relative abundances of IgG N-glycans. Please click here to view a larger version of this figure.
Supplementary Figure 1: The gradient program applied in the study is illustrated. Please click here to download this File.
Supplementary Figure 2. Mass spectrometric parameters applied in the study. Please click here to download this File.
Supplementary Figure 3: The parameters for searching glycan structures used in the study. Please click here to download this File.
Supplementary Figure 4: Parameters applied for the extraction of peak areas. Please click here to download this File.
Supplementary Figure 5. Annotated MS/MS spectra of IgG N-glycans. (A) H3N5F1 (B) H4N4F1. Please click here to download this File.
Supplementary Table 1: The list of N-glycans belonging to IgG obtained from HILIC-FLD-MS/MS analysis. Please click here to download this Table.
Supplementary Table 2: The list of N-glycans belonging to trastuzumab obtained from HILIC-FLD-MS/MS analysis. Please click here to download this Table.
Supplementary Table 3: The list of N-glycans belonging to human plasma glycoproteome obtained from HILIC-FLD-MS/MS analysis. Please click here to download this Table.
Discussion
N-glycan profiling of glycoproteins includes challenging steps. Although there are many different methodologies for this purpose, a suitable approach should be selected for both identification and quantification of N-glycan structures14. HILIC-FLD is the gold standard approach for the quantification of N-glycans. However, identification of all N-glycan types by FLD detection is not achieved. Therefore, tandem MS analysis is needed for confirming N-glycan structures derived from glycoproteins. By the combination of FLD and MS detection in the same system, analysis of the N-glycans is more efficient23. Due to these reasons, both identification and quantification of N-glycans are performed by HILIC-FLD-MS/MS for standard glycoproteins as well as complex samples such as human plasma.
N-glycans are labeled by a fluorescence tag from their reducing ends to detect and quantify them by FLD25. N-glycans have low ionization efficiencies in mass spectrometric analysis. Procainamide is a fluorescence tag that improves the ionization efficiencies compared with commonly applied tags such as 2-AB (2-aminobenzamide)20,21. Thus, we selected this tag for achieving tandem MS analysis of N-glycans efficiently. This tag also allows confirmation of core fucosylated and bisected N-glycan types by monitoring specific fragments obtained from tandem MS analysis22,23 as stated in the presented protocol.
Collision-induced dissociation (CID) or higher-energy C-trap dissociation (HCD) is commonly applied to interpret the N-glycan spectra8. The MS/MS spectra can allow detection of the branching points and elongation of the N-glycan structure26. In addition, the linkage position of N-glycans can be assigned by monitoring cross-ring fragmentation in CID and HCD27. However, data analysis software is necessary to identify N-glycan structures together with instrumentation and bioanalytical methods28. Several commercially or freely available tools from literature can be used with the presented approach for interpreting N-glycan structures.
Purification of labeled N-glycans is usually applied before HILIC-FLD-MS/MS analysis because sample preparation steps contain various chemicals that have interfered with the analysis. Several commercially available sorbents based on HILIC interaction can be used for the purification of N-glycans29. Cellulose is one of the cheapest alternatives for the purification of N-glycans. It is employed not only for batch-mode experiments but also for solid-phase extraction applications. In addition, purifications can be achieved in 96 well plate platforms using cellulose. The presented method with the purification step may be optimized depending on the sample size. On the other hand, alternative materials such as porous graphitized carbon (PGC) can be inserted into the protocol to purify procainamide labeled N-glycans. Furthermore, PGC-based purification can be used with cellulose-based HILIC purification to increase purification efficiency30.
Data analysis for profiling N-glycans is a time-consuming process. Recently, a python-based open-source tool to quantify peaks of N-glycans has been demonstrated24. This tool allows automated data analysis workflow for peak selection and retention time calibration. In addition, a large amount of data belonging to N-glycan samples can be analyzed by a batch-mode application. This tool makes the data analysis faster and easier. Additionally, other software can be inserted into the current strategy to evaluate the obtained data. In conclusion, a facile method was demonstrated here to profile N-glycans using HILIC-FLD-MS/MS. This approach can apply to any glycoproteins such as protein biopharmaceuticals as well as complex samples to profile N-glycans.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was partly supported by the Ministry of Development-Republic of Turkey with project number: 2016 K121230. Bekir Salih gratefully acknowledges the Turkish Academy of Science (TUBA) for the partial financial support.
Materials
| Acetic acid | Carlo Erba Reagents | 401413 | Glacial RS For LC/MS |
| Acetonitrile | Merck | 1000292500 | LC-MS LiChrosolv |
| Agilent 1200 Series HPLC with 1260 Series FLD dedector | Agilent Technologies | ||
| Ammoniumm Formate | Carlo Erba Reagents | 419741 | For LC/MS |
| Bruker TIMS-TOF (Q-TOF) Mass Spectrometry | Bruker Daltonics | ||
| Cellulose | Sigma Aldrich | 310697 | microcrystalline, powder, 20 μm |
| Deionized Water | Carlo Erba Reagents | 412111 | For LC/MS |
| Dimethyl sulfoxide | Sigma Aldrich | 41639 | BioUltra, for molecular biology, ≥99.5% (GC) |
| Empty polypropylene SPE Tube with PE frits | Sigma Aldrich | 54220 | 20 μm porosity,volume 1 mL |
| Extraction Manifold, 20-position | Waters | WAT200607 | Complete with rack for 13 x 100 mm tubes |
| Human Plasma | Sigma Aldrich | P9523 | lyophilized |
| IGEPAL CA-630 | Sigma Aldrich | I8896 | for molecular biology |
| IgG | Sigma Aldrich | I4506 | lyophilized powder |
| Phosphate buffered saline | Sigma Aldrich | P4417 | Tablet |
| PNGase F enzyme | Promega | V483A | |
| Procainamide hydrochloride | abcam | ab120955 | |
| Sodium cyanoborohydride | Sigma Aldrich | 156159 | reagent grade, 95% |
| Sodium dodecyl sulfate | Sigma Aldrich | 71725 | |
| trastuzumab | Roche Diagnostics | ||
| Trifluoroacetic acid | Sigma Aldrich | 302031 | for HPLC, ≥99.9% |
References
- Dwek, R. A. Glycobiology: Toward understanding the function of sugars. Chemical Reviews. 96 (2), 683-720 (1996).
- Varki, A. Biological roles of glycans. Glycobiology. 27 (1), 3-49 (2016).
- Spiro, R. G. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 12 (4), 43 (2002).
- Apweiler, R., Hermjakob, H., Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica et Biophysica Acta. 1473 (1), 4-8 (1999).
- Stavenhagen, K., et al. and O-glycosylation Analysis of Human C1-inhibitor Reveals Extensive Mucin-type O-Glycosylation. Molecular & Cellular Proteomics. 17 (6), 1225-1238 (2018).
- Ząbczyńska, M., Kozłowska, K., Pocheć, E. Glycosylation in the Thyroid Gland: Vital Aspects of Glycoprotein Function in Thyrocyte Physiology and Thyroid Disorders. International Journal of Molecular Sciences. 19 (9), (2018).
- Mallet, B., et al. N-Glycans Modulate in Vivo and in Vitro Thyroid Hormone Synthesis: Study at the N-Terminal Domain Of Thyroglobulin. Journal of Biological Chemistry. 270 (50), 29881-29888 (1995).
- Dong, X., et al. Advances in mass spectrometry-based glycomics. Electrophoresis. 39 (24), 3063-3081 (2018).
- Ohtsubo, K., Marth, J. D. Glycosylation in cellular mechanisms of health and disease. Cell. 126 (5), 855-867 (2006).
- Koçak, &. #. 2. 1. 4. ;. F., et al. N-glycan profiling of papillary thyroid carcinoma tissues by MALDI-TOF-MS. Analytical Biochemistry. 584, 113389 (2019).
- Peng, W., et al. Clinical application of quantitative glycomics. Expert Review of Proteomics. 15 (12), 1007-1031 (2018).
- Yaman, M. E., Kayili, H. M., Albayrak, M., Kadioglu, Y., Salih, B. Differential N-glycosylation profiling of formalin-fixed paraffin-embedded (FFPE) invasive ductal carcinoma tissues using MALDI-TOF-MS. Molecular Omics. 17 (3), 394-404 (2021).
- Liu, L. Antibody Glycosylation and Its Impact on the Pharmacokinetics and Pharmacodynamics of Monoclonal Antibodies and Fc-Fusion Proteins. Journal of Pharmaceutical Sciences. 104 (6), 1866-1884 (2015).
- Zhang, L., Luo, S., Zhang, B. Glycan analysis of therapeutic glycoproteins. mAbs. 8 (2), 205-215 (2016).
- Hart, G. W., Copeland, R. J. Glycomics Hits the Big Time. Cell. 143 (5), 672-676 (2010).
- West, C. M., Malzl, D., Hykollari, A., Wilson, I. B. H. Glycomics, Glycoproteomics, Glycogenomics: An Inter-Taxa Evolutionary Perspective. Molecular & Cellular Proteomics. 20, 100024 (2021).
- Unione, L., et al. Glycoprofile Analysis of an Intact Glycoprotein As Inferred by NMR Spectroscopy. ACS Central Science. 5 (9), 1554-1561 (2019).
- Morelle, W., Michalski, J. -. C. Analysis of protein glycosylation by mass spectrometry. Nature Protocols. 2 (7), 1585-1602 (2007).
- Reusch, D., et al. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles–part 1: separation-based methods. mAbs. 7 (1), 167-179 (2015).
- Keser, T., Pavić, T., Lauc, G., Gornik, O. Comparison of 2-Aminobenzamide, Procainamide and RapiFluor-MS as Derivatizing Agents for High-Throughput HILIC-UPLC-FLR-MS N-glycan Analysis. Frontiers in Chemistry. 6, 324 (2018).
- Kozak, R. P., Tortosa, C. B., Fernandes, D. L., Spencer, D. I. R. Comparison of procainamide and 2-aminobenzamide labeling for profiling and identification of glycans by liquid chromatography with fluorescence detection coupled to electrospray ionization-mass spectrometry. Analytical Biochemistry. 486, 38-40 (2015).
- Nwosu, C., Yau, H. K., Becht, S. Assignment of Core versus Antenna Fucosylation Types in Protein N-Glycosylation via Procainamide Labeling and Tandem Mass Spectrometry. Analytical Chemistry. 87 (12), 5905-5913 (2015).
- Kayili, H. M. Identification of bisecting N-glycans in tandem mass spectra using a procainamide labeling approach for in-depth N-glycan profiling of biological samples. International Journal of Mass Spectrometry. 457, 116412 (2020).
- Jansen, B. C., et al. HappyTools: A software for high-throughput HPLC data processing and quantitation. PLOS ONE. 13 (7), 0200280 (2018).
- Ruhaak, L. R., et al. Glycan labeling strategies and their use in identification and quantification. Analytical and bioanalytical chemistry. 397 (8), 3457-3481 (2010).
- Rojas-Macias, M. A., et al. Towards a standardized bioinformatics infrastructure for N- and O-glycomics. Nature Communications. 10 (1), 3275 (2019).
- Everest-Dass, A. V., Abrahams, J. L., Kolarich, D., Packer, N. H., Campbell, M. P. Structural feature ions for distinguishing N- and O-linked glycan isomers by LC-ESI-IT MS/MS. Journal of the American Society for Mass Spectrometry. 24 (6), 895-906 (2013).
- Li, X., Xu, Z., Hong, X., Zhang, Y., Zou, X. Databases and Bioinformatic Tools for Glycobiology and Glycoproteomics. International Journal of Molecular Sciences. 21 (18), 6727 (2020).
- Qing, G., Yan, J., He, X., Li, X., Liang, X. Recent advances in hydrophilic interaction liquid interaction chromatography materials for glycopeptide enrichment and glycan separation. TrAC Trends in Analytical Chemistry. 124, 115570 (2020).
- Reiding, K. R., et al. Human Plasma N-glycosylation as Analyzed by Matrix-Assisted Laser Desorption/Ionization-Fourier Transform Ion Cyclotron Resonance-MS Associates with Markers of Inflammation and Metabolic Health. Molecular & Cellular Proteomics. 16 (2), 228-242 (2017).

.
