TGF-β-mediated Endothelial to Mesenchymal Transition (EndMT) and the Functional Assessment of EndMT Effectors using CRISPR/Cas9 Gene Editing
Summary
We describe methods to investigate TGF-β2-induced EndMT in endothelial cells by observing cell morphology changes and examining the expression EndMT-related marker changes using immunofluorescence staining. CRISPR/Cas9 gene editing was described and used to deplete the gene encoding Snail to investigate its role in TGF-β2-induced EndMT.
Abstract
In response to specific external cues and the activation of certain transcription factors, endothelial cells can differentiate into a mesenchymal-like phenotype, a process that is termed endothelial to mesenchymal transition (EndMT). Emerging results have suggested that EndMT is causally linked to multiple human diseases, such as fibrosis and cancer. In addition, endothelial-derived mesenchymal cells may be applied in tissue regeneration procedures, as they can be further differentiated into various cell types (e.g., osteoblasts and chondrocytes). Thus, the selective manipulation of EndMT may have clinical potential. Like epithelial-mesenchymal transition (EMT), EndMT can be strongly induced by the secreted cytokine transforming growth factor-beta (TGF-β), which stimulates the expression of so-called EndMT transcription factors (EndMT-TFs), including Snail and Slug. These EndMT-TFs then up- and downregulate the levels of mesenchymal and endothelial proteins, respectively. Here, we describe methods to investigate TGF-β-induced EndMT in vitro, including a protocol to study the role of particular TFs in TGF-β-induced EndMT. Using these techniques, we provide evidence that TGF-β2 stimulates EndMT in murine pancreatic microvascular endothelial cells (MS-1 cells), and that the genetic depletion of Snail using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated gene editing, abrogates this phenomenon. This approach may serve as a model to interrogate potential modulators of endothelial biology, and can be used to perform genetic or pharmacological screens in order to identify novel regulators of EndMT, with potential application in human disease.
Introduction
Endothelial to mesenchymal transition (EndMT) is a multistep and dynamic biological phenomenon that has been linked to diverse physiological and pathological processes1,2. Upon EndMT endothelial cells gradually lose their endothelial traits, while acquiring mesenchymal properties3; thus, tightly compacted and well organized endothelial cells differentiate into elongated mesenchymal-like cells. Morphological changes in EndMT coincide with alterations in the expression of certain genes and proteins. In general, the expression of proteins that maintain endothelial characteristics, including vascular endothelial (VE)-cadherin, platelet/EC adhesion molecule-1 (CD31/Pecam-1) declines. Simultaneously, proteins related to mesenchymal functions, such as α-smooth muscle actin (α-Sma) and smooth muscle protein 22α (Sm22α) accumulate. Emerging results have demonstrated that postnatal EndMT contributes to the development of human diseases, such as cancer, cardiac fibrosis, pulmonary arterial hypertension (PAH), atherosclerosis (AS), organ fibrosis, etc2,4,5,6,7. A deeper understanding of the underlying mechanisms of EndMT and how to direct the EndMT process will provide novel therapeutic methods for EndMT-related diseases and regenerative medicine.
TGF-β is one of the main EndMT inducers, and other known involved factors include Wnt/β-catenin, Notch, and some inflammatory cytokines1. As the cellular context is key for responses triggered by TGF-β, the interplay of TGF-β with other EndMT promoting signals is relevant for TGF-β to elicit an EndMT response. Upon the activation of TGF-β cell surface type I and type II serine/threonine kinase receptors, the intracellular canonical Smad pathway is activated. TGF-β receptor-mediated phosphorylated Smad2/3 form heteromeric complexes with Smad4 that translocate into the nucleus, where they upregulate the expression of EndMT-related transcription factors. Similar to epithelial-mesenchymal transition (EMT), transcription factors such as Snail, Slug, Twist, Zeb1 and Zeb2 are induced by TGF-β signaling and contribute to gene reprogramming in EndMT8.
Snail has been frequently identified as a key factor in EndMT. Snail binds to the promoter of genes encoding cell-cell adhesion proteins and suppresses their transcription, which is counterbalanced by the enhancement of the expression of mesenchymal proteins9. Endothelial cells comprise a very heterogeneous population and the relative influence of diverse extracellular stimuli on EndMT may differ among endothelial cellular contexts or cell types10. Due to its similarities with EMT, some methodologies are useful to investigate both mechanisms EMT and EndMT8. In this regard, the EMT International Association (TEMTIA) strongly emphasizes the need of complementary techniques to ultimately demonstrate the occurrence of EMT/EndMT11.
Here we describe a method to monitor and visualize the TGF-β-induced EndMT process. Immunofluorescence staining provides the basic information about expression changes in targeted proteins/markers, which are used as indicators of whether the EndMT process occurs. Additionally, the immunofluorescence staining can visualize the localization of proteins/markers and cell morphology. To study the potential activity of specific TFs (or other upstream or downstream regulators) involved in TGF-β mediated EndMT, we describe a protocol using clustered regularly interspaced short palindromic repeats (CRISPR) /CRISPR-associated protein 9 (Cas9) gene editing to deplete specific genes from cells, using the TF Snail as an example. Cas9 is a dual RNA-guided DNA endonuclease that recognizes and cleaves sequences complementary to CRISPR sequences in bacteria12. The CRISPR/Cas9 system is currently extensively utilized because it facilitates genetic engineering in vitro and in vivo13. Directed by a single guide RNA (sgRNA), ectopically expressed Cas9 generates a double strand break at a preselected targeting sequence in a specific gene locus. Non-homologous end joining (NHEJ) takes place to repair Cas9-induced strand breaks, via random nucleotide insertions or deletions thereby leading to the disruption and inactivation of the targeted gene. We describe in detail methods for designing selective sgRNAs and generating lentiviral-compatible vectors containing the designed sgRNAs. As a result, stable gene-depleted endothelial cells can be generated in an efficient and reliable manner.
In this study, we used murine pancreatic microvascular endothelial cells (MS-1)14 as a model system to examine the TGF-β2-induced EndMT process. Our previous study demonstrated that Snail is the main transcription factor increased by TGF-β2, by which EndMT is induced in MS-1 cells15. Upon CRISPR/Cas9 gene editing to abrogate Snail expression in MS-1 cells, TGF-β2 failed to mediate EndMT. This workflow can be applied to study other (suspected) EndMT-related genes.
Protocol
1. Induction of EndMT by TGF-β2
- MS-1 cells in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin in an incubator (5% CO2, 37 °C). Coat all culture dishes/plates with 0.1% w/v gelatin for 10 min before use.
- Gently wash MS-1 cells with 1x phosphate buffered saline (PBS), add 2 mL of trypsin-EDTA solution (0.25% trypsin and 0.02% EDTA) to a 10 cm dish, and incubate for 2 min at 37 °C to detach them. Subsequently, add 5 mL of complete culture medium to quench the reaction.
- Transfer the cell suspension to a 15 mL tube and centrifuge at 200 x g for 3 min at room temperature.
- Discard the supernatant and resuspend the cells in 4 mL of fresh medium containing FBS and penicillin/streptomycin. Count the cells using an automatic cell counter.
- Seed 1 x 103 cells per cm2 for further culture. For example, seed 9.5 x 103 cells/well for 6-well plates, or 1.9 x 103 cells/well for 24-well plates.
- Incubate the cells overnight to allow them to adhere and recover, then stimulate MS-1 cells with TGF-β2 for 3 days. Add the TGF-β receptor kinase inhibitor SB431542 (5 µM) 30 min before TGF-β2 stimulation. Treat other cells with vehicle (DMSO).
NOTE: Dissolve TGF-β2 in 4 mM HCl containing 0.1% human bovine serum albumin (BSA). Add the same amount of ligand buffer without TGF-β2 to the control group. The TGF-β2 concentration may be adapted to be 0.1-1 ng/mL for specific assays. See indications in the corresponding figures. - After 3 days, examine the cell morphology with bright field imaging (with an inverted microscope) and perform immunofluorescence staining (see step 2) to assess EndMT-related marker changes. Perform at least three independent experiments to obtain biological triplicates.
2. Immunofluorescence staining
- Trypsinize cultured MS-1 cells (step 1.2) and then reseed 1.9 x 103 cells on a 0.1% w/v gelatin-coated 12 mm round cover glass placed on the bottom of a 24-well plate.
- After culturing the cells overnight, add TGF-β2 (final concentration 1 ng/mL) to the cells for 3 days. Use medium containing ligand buffer as a negative control.
- Perform Pecam-1 and Sm22α staining.
- After stimulating the cells with TGF-β2 (or Control) for 3 days, remove the medium, and wash the cells with 1x PBS.
- Add 300 µL of 4% formaldehyde to each well and incubate for 10 min at room temperature to fix the cells. Wash 3x with 1x PBS after incubation.
- Add 300 µL of 0.1% Triton X-100 in 1x PBS to each well and incubate for 10 min at room temperature to permeabilize the cells. After incubation, remove the Triton X-100 solution and wash the cells 3x with 1x PBS.
- Block the cells with 3% bovine serum albumin (BSA) in 1x PBS for 45 min at room temperature.
- Dilute the primary Pecam-1 and Sm22α antibodies that recognize murine proteins 1:500 with 1x PBS. Then incubate the fixed cells with primary antibodies for 45 min at room temperature.
- After washing 3x with 1x PBS, incubate the cells with 1000x diluted secondary antibodies, including donkey anti-rat Alexa 488 and goat anti-rabbit Alexa 594, for 45 min at room temperature.
NOTE: Protect the samples from light during staining. - After rinsing 3x with 1x PBS, place the cover glass seeded with cells face down on a drop of mounting medium containing 4',6-diamidino-2-phenylindole (DAPI) on a slide to stain the nuclei.
- Fix the periphery of the cover glass with transparent nail polish and store it at 4 °C.
- Acquire representative images with a confocal microscope. Set the laser wavelengths at 405 nm, 488 nm and 552 nm to detect DAPI, Pecam-1 and Sm22α, respectively. For each channel, all the pictures were taken with the same settings and exposure time. Perform at least three independent experiments to obtain biological triplicates.
3. Knock out of Snail using CRISPR/Cas9 editing
- Design two independent sgRNAs targeting murine Snail.
- Design sgRNAs using the online tools CHOPCHOP (https://chopchop.cbu.uib.no/) and Cas-OFFinder (http://www.rgenome.net/cas-offinder/) according to the targeted gene name and species.
- Predict the off-target activity of the designed sgRNAs targeting to Snail with two independent algorithms, including Cas-OFFinder (http://www.rgenome.net/cas-offinder/) and CHOPCHOP (http://chopchop.cbu.uib.no/).
- Choose two sgRNAs with the lowest off-activity. Design two complementary sgRNA oligos with the BveI cut site. The sense oligo starts with 5'-ACCG-3' and the antisense oligo starts with 5'-AAAC-3'.
- Order the oligos to be commercially synthesized for further use.
- Clone the complementary guide RNA oligos into the BveI-digested AA19 pLKO.1-puro.U6.sgRNA.BveI-stuffer Lentiviral vector plasmid to generateAA19 pLKO.1-Snail-sgRNA16.
- Cut the AA19 pLKO.1-puro.U6.sgRNA.BveI-stuffer plasmid with the BveI enzyme16. Mix 2 µg of AA19 pLKO.1-puro.U6.sgRNA.BveI-stuffer plasmid, 5 µL of 10x Buffer O, and 5 µL of BveI enzyme and add sterile water to reach a total volume of 50 µL.
- Vortex and briefly spin down the reaction mix. Incubate the reaction at 37 °C for 1 h.
- Load the reaction mix on a 1% agarose gel and run in 1x Tris-acetate-EDTA (TAE, 50x TAE stock: 242 g of Tris base dissolved in water, 57.1 mL of glacial acetic acid, 100 mL of 500 mM EDTA (pH 8.0) solution, and add water to a total of 1 L) buffer until a good separation is achieved.
- Cut the backbone fragment from the gel, isolate it with a gel extraction kit according to the manufacturer's protocol (see the Table of Materials) and elute the backbone in 40 µL elution buffer (EB).
- Mix 5 µL of 100 pmol/µL sense oligo and 5 µL of 100 pmol/µL antisense oligo with 1 µL of 1 M Tris-HCl (pH 8.0) and add sterile water to reach a total of 100 µL to anneal the gRNA oligos. Incubate the mixture for 5 min at 100 °C, and then cover the tubes with aluminum foil. After that, slowly cool the solution to room temperature for further use as an insert.
- To ligate the complementary gRNA oligos and the BveI-digested backbone, mix 1 µL of isolated BveI cut AA19 pLKO.1-puro.U6.sgRNA.BveI-stuffer backbone and 2 µL of insert (diluted 1:300) with 2 µL of 10x T4 DNA ligase buffer and 1 µL of T4 DNA ligase, and add sterile water to reach a total of 20 µL. Briefly spin the tube, and incubate it for 4 h at room temperature for further use.
NOTE: When performing the ligation, ensure two control groups are included. For one control group, mix 1 µL of isolated backbone, 2 µL of 10x T4 DNA ligase buffer, and 1 µL of T4 DNA ligase and add sterile water to reach a total of 20 µL, but without oligo DNA. For the other control group, mix 1 µL of isolated backbone and 2 µL of 10x T4 DNA ligase buffer, and add sterile water to reach a total of 20 µL, but without the annealed oligos and T4 ligase. These two control ligations are used to determine the reaction background in the next transformation step and indicate of how efficient the ligation is.
- Transform the reaction mixture into competent TOP10 E. coli.
- Collect the competent TOP10 E. coli. cells from the -80 °C freezer, and thaw them on ice.
- Add 2 µL of ligation mixture to 50 µL of competent cells and keep the tube on ice for 30 min.
- Heat-shock the tube at 42 °C for 30 s. Put the tube on ice for 2 min.
- Add 950 µL of fresh lysogeny broth (LB) medium to the mixture and shake vigorously at 37 °C for 60 min.
- Spin down and plate the cells on a warm ampicillin (100 µg/mL) resistance LB plate. Incubate the plate at 37 °C overnight.
- Verify the successful insertion of the gRNA oligos in the plasmid.
- Pick 3-5 colonies on the plate in 1 mL of ampicillin (100 µg/mL) containing LB medium and shake overnight at 30 °C.
- Isolate the plasmid DNA with a plasmid kit according to the manufacturer's protocol (Table of Materials) and sequence it with the U6 promoter primer 5'- GAGGGCCTATTTCCCATGATT -3' to verify the successful insertion of the gRNA oligo.
4. Generate Snail knockout MS-1 cells
- Produce lentiviral particles carrying Cas9 or Snail-targeting gRNAs.
- Culture HEK 293T cells in DMEM containing 10% fetal bovine serum and 100 U/mL penicillin/streptomycin in 14.5 cm dishes (or T75 flasks) in an incubator (5% CO2, 37 °C).
- Mix 9.9 µg of targeting gene plasmid, AA19 pLKO.1-Snail-sgRNA or pLV-Cas917 plasmid, together with the helper plasmids 3.5 µg of pCMV-VSVG (encoding the G protein of the vesicular stomatitis virus, VSV-G), 6.6 µg of rev-responsive element plasmid pMDLg-RRE (encoding Gag and Pol), and 5.0 µg of pRSV-REV (encoding Rev) in 500 µL of serum free medium. Resuspend 50 µL of polyethylenimine (PEI) (2.5 mg/mL) in 500 µL of serum free medium. Gently mix plasmids and PEI preparations by pipetting up and down. Incubate the mixture for 20 min at room temperature.
- Transfect HEK 293T cells by adding the mixture medium from step 4.1.2 to 80% confluent cells in 14.5 cm dishes (or T75 flasks) which contain DMEM medium with 10% FBS and 100 U/mL penicillin/streptomycin. HEK293T cells are used because they are easily transfected and generate high levels of virus18.
- Transfer transfected HEK 293T cells to a biosafety in microbiological and biomedical laboratory (BMBL) to culture them for 24 h.
- In a BMBL laboratory, replace the transfection medium from HEK 293T cells with 12 mL of fresh complete DMEM containing FBS and penicillin/streptomycin. Incubate the cells for 24 h.
- Collect and filter the medium with a 20 mL syringe and a 0.45 µm filter. Transfer the conditioned medium into a 15 mL polypropylene tube.
- Add 12 mL of fresh complete DMEM containing FBS and penicillin/streptomycin to the HEK 293T culture dish and culture for an additional 24 h.
- Collect and filter the medium with a 20 mL syringe and a 0.45 µm filter. Transfer the conditioned medium into a 15 mL polypropylene tube. Store the medium containing lentiviral particles as 1 mL aliquots at -80 °C for further use.
- Infect MS-1 cells with the pLV-Cas9 virus.
- Seed 1 x 105 MS-1 cells per well in a 6-well plate for 24 h before lentivirus infection.
- Thaw the frozen aliquots of pLV-Cas9 virus in a 37 °C water bath.
- Mix 1 mL of virus medium with 1 mL of fresh DMEM medium containing FBS and penicillin/streptomycin. Add polybrene to the medium (final concentration as 10 μg/mL) to increase the infection efficiency.
- Remove the medium from the 6-well plate and replace it with the virus/polybrene mix medium and culture the cells in an incubator for 24 h. 24 h post-infection, replace the medium with fresh medium and culture the cells for another 24 h.
NOTE: Always keep an uninfected well as a control group. - Aspirate the medium from the infected group and control group and replace it with DMEM medium with 4 µg/mL blasticidin.
- Return the plate to a 37 °C incubator and culture the cells for 1 week. Uninfected cells will die due to the effect of blasticidin. Split the surviving cells when they reach 80% cell confluency and continue with blasticidin selection.
- Confirm the expression of Cas9 in MS-1 cells by western blotting using an antibody against Cas9 (molecular weight of Cas9 is approximately 160 kDa).
- Separately infect pLV-Cas9 MS-1 cells with two independent gRNA lentiviruses.
- Seed 1 x 105 pLV-Cas9 MS-1 cells per well in a 6-well plate for 24 h before infection.
- Follow the same protocol as described in 4.2 to separately infect cells with two gRNA lentiviruses.
- After 24 h of infection with the gRNA virus, refresh the medium and culture the cells for another 24 h.
- Replace the medium with DMEM with 1 µg/mL puromycin. Return the plate to a 37 °C incubator and culture the cells for 1 week. Ensure the uninfected cells are completely dead. Split the cells when they reach 80% confluency and continue puromycin selection.
- Confirm the knock-out of Snail in MS-1 cells by western blotting using an antibody against Snail (molecular weight of Snail is approximately 35 kDa).
Representative Results
TGF-β2 induces EndMT and stimulates Snail expression in MS-1 endothelial cells
TGF-β is one of the cytokines with greatest potential to induce EndMT. After treating MS-1 cells with TGF-β2 (1 ng/mL) for 3 days, endothelial MS-1 cells lose their cobblestone-like structure and differentiate into spindle-shaped mesenchymal-like cells (Figure 1A)15. To further verify the role of TGF-β2 in inducing cell phenotypic changes, we pre-treated the cells with the small molecule activin receptor-like kinase (ALK)4/ALK5/ALK7 inhibitor SB431542 before TGF-β2 stimulation19. SB431542 completely abrogated TGF-β2-induced cell morphology changes (Figure 1A). The TGF-β2 induced EndMT process was further investigated by studying changes in the expression of EndMT-related markers. As shown in Figure 1B, the endothelial protein Pecam-1 was potently decreased after TGF-β2 stimulation, while the mesenchymal factor Sm22α was profoundly upregulated by TGF-β215. These data are consistent with the notion that TGF-β2 triggered EndMT in MS-1 cells. Next, we investigated the effects of TGF-β2 on Snail and Slug expression. As shown in Figure 1C, Snail was markedly upregulated by TGF-β2, while Slug expression was not influenced by TGF-β2 in MS-1 cells15. The quantification of Snail expression from three independent experiments is shown in Figure 1D.
Depletion of Snail by CRISPR/Cas9 in MS-1 endothelial cells
As Snail was induced by TGF-β2 and likely involved in TGF-β2-mediated EndMT, we performed CRISPR/Cas9 gene editing to genetically deplete Snail expression in MS-1 cells. We hypothesized that the depletion of Snail would be sufficient to inhibit TGF-β2-induced EndMT. As shown in Figure 2A, we generated Snail knockout cells in two steps. Firstly, Cas9 was ectopically expressed by infecting MS-1 cells with a Cas9 expressing lentivirus. Since there is a blasticidin resistance cassette in the pLV-Cas9 construct, we checked the expression of Cas9 by Western blot analysis in blasticidin resistant cells (Figure 2D). Subsequently, we introduced sgRNAs that specifically targeted Snail to disrupt its protein expression. This procedure was also performed by infection with lentiviral particles carrying the AA19 pLKO.1-Snail-sgRNA construct, which includes a puromycin expression cassette. Cas9-expressing cells were again infected with gRNA containing lentivirus and further selected with puromycin. Two complementary sgRNA oligos targeting murine Snail were designed with a predicted low off-target activity (Figure 2B,C). After introducing two independent Snail sgRNAs in Cas9 expressing MS-1 cells, Snail protein expression was abrogated (Figure 2D)15.
Deficiency of Snail inhibits TGF-β2-induced EndMT in MS-1 cells
To demonstrate the function of Snail in TGF-β2-mediated EndMT, we performed an EndMT assay in Snail-depleted cells and compared it with parental MS-1 cells. As shown in Figure 3A, the knockout of Snail was sufficient to inhibit the fibroblast-like cell morphology driven by TGF-β2 in MS-1 cells15. In addition, the TGF-β2-mediated decline in Pecam-1 and enhancement of Sm22α were completely blocked in Snail-depleted MS-1 cells. In summary, we demonstrated that Snail is critical for TGF-β2-mediated EndMT in MS-1 cells (Figure 3B)15.
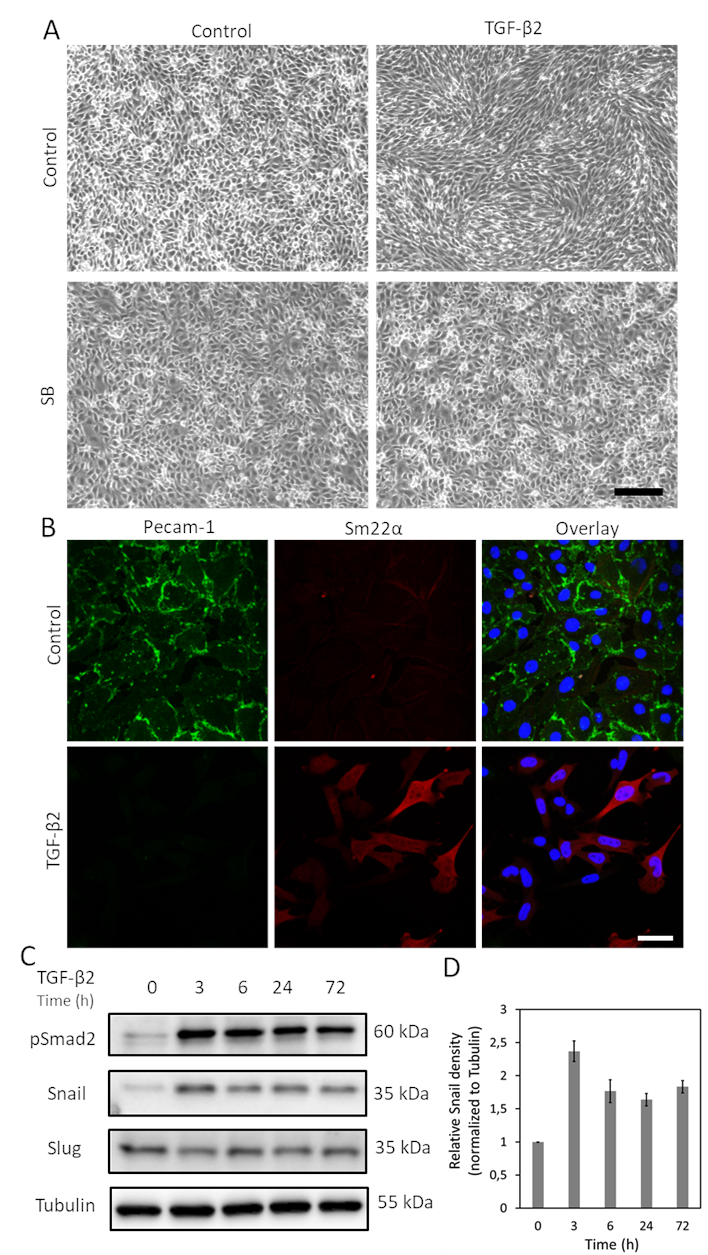
Figure 1. TGF-β2 induces EndMT and Snail expression in MS-1 cells. A. Effects of TGF-β2 and/or TGF-β type I receptor kinase inhibitor SB-431542 on cell morphology. Brightfield images of MS-1 cells upon treatment with TGF-β2 (1 ng/mL) and/or SB-431542 (SB, 5 µM, administered 30 min prior to TGF-β2) for 2 days. Scale bar represents 200 µm. B. Immunofluorescence staining of Pecam-1 (green) and Sm22α (red) in MS-1 cells cultured in medium containing TGF-β2 (1 ng/mL) for 3 days. Nuclei are visualized in blue (DAPI). Scale bar: 50 µm. C. Western blot with whole cell lysate of TGF-β2 stimulated MS-1 cells. The expression of Snail, but not Slug, was enhanced by TGF-β2 stimulation, as previously reported in Ma et al15. D. Quantification of Snail expression by integrating the results from three independent western blot experiments. Please click here to view a larger version of this figure.

Figure 2. Depletion of Snail by CRISPR-Cas9 gene editing. A. Scheme depicting how to generate Snail knockout cells. Bsd: Blasticidin. Puro: Puromycin. B. Oligonucleotides of two independent sgRNAs targeting Snail using CHOPCHOP (http://chopchop.cbu.uib.no/) and Cas-OFFinder (http://www.rgenome.net/cas-offinder/). C. The predicted off-target activity of the two gRNAs for Snail using Cas-OFFinder (http://www.rgenome.net/cas-offinder/). D. Cas9 and Snail expression in wild type (WT) and Cas9-overexpressed MS-1 measured by Western blot analysis. E. Knockout of Snail with two independent gRNAs in MS-1 cells as measured by Western blot analysis. We reported similar results in Ma et al15. Please click here to view a larger version of this figure.

Figure 3. Genetic depletion of Snail inhibits TGF-β2-induced EndMT in MS-1 cells. A. Brightfield images of MS-1 cells upon treatment with TGF-β2 (0.1 ng/mL) for 3 days in wildtype (WT, upper panel) and Snail knocked out (lower panel) cells. Scale bar represents 200 µm. B. Immunofluorescent staining for Pecam-1 (green), Sm22α (red) and nuclei (blue) of MS-1 cells cultured in medium containing TGF-β2 (1 ng/mL) for 3 days. Depletion of Snail abrogated TGF-β2-induced decrease of Pecam-1 and increase of Sm22α expression. Scale bar represents 50 µm. C. Schematic representation of the effect of Snail knockout on TGF-β-induced EndMT in MS-1 cells. TGF-β stimulates the expression of Snail through Smad pathway by phosphorylating Smad2/3 and further drives EndMT. Knocking out Snail using CRISPR/Cas9-based gene editing abrogated TGF-β-mediated EndMT. Please click here to view a larger version of this figure.
Discussion
Understanding the mechanism of EndMT is critical for modulating this process and targeting EndMT-related diseases. Here, we described methods to perform a TGF-β-induced EndMT assay and interrogate the role of the EndMT-TF Snail in TGF-β-triggered EndMT, by performing CRISPR/Cas9-mediated stable gene depletion of Snail from cells. The depletion of Snail using CRISPR/Cas9 approach successfully abrogated TGF-β2 driven EndMT in MS-1 cells (Figure 3C). To study the effects of any cytokines, like TGF-β, on EndMT, ECs were exposed to cytokines and then the occurrence of EndMT was assessed according to morphological changes and endothelial and mesenchymal marker expression changes in cells. TGF-β2 strongly induced EndMT in MS-1 cells accompanied by a strong increase in the expression of the transcription factor Snail. The EndMT-TFs induced by TGF-β can differ according to the species or tissue-specific endothelial cell type. For example, we observed that Snail but not Slug was significantly upregulated by TGF-β in MS-1 cells, while in human umbilical vein endothelial cells (HUVECs), both Snail and Slug are increased after exposure to TGF-β20.
We assessed the extent of the EndMT process in two ways by examining cell morphology changes and then by investigating changes in EndMT-related markers expression. After TGF-β exposure for 3 days, cells underwent EndMT with consistent morphological variations and changes in the expression of EndMT-related markers. In addition to the immunofluorescence staining we performed here, marker variations can also be monitored by western blotting at the protein expression level or by qRT-PCR (Real-Time Quantitative Reverse Transcription PCR) at the gene levels21. In addition to these two time- and cost-saving methods that we showed in this protocol, there are other methods to examine EndMT. For example, performing transcriptome analysis (by RNA sequencing or qPCR) to compare the expression levels of endothelial- and mesenchymal-related genes between treated and control cells can precisely assess EndMT22,23. In addition, EndMT often involves the stable loss of barrier function, which can be assessed by impedance spectroscopy24. Furthermore, additional proof of the acquisition of stem cell-like properties by EndMT-derived cells may be examined. For instance, under specific culture conditions, EndMT mesenchymal-like cells can be further differentiated into osteoblasts, chondrocytes, adipocytes or (myo)fibroblasts. Therefore, additional analysis to confirm the differentiation into different cell types belonging to the mesoderm lineage (i.e., gene expression and matrix staining) is useful for demonstrating the multipotent nature of EndMT-derived cells. Finally, the EndMT assessment methods are not limited to in vitro studies, but can be extrapolated to investigate the relation between EndMT and some diseases in vivo or in ex vivo organs. In this sense, the use of endothelial-specific lineage tracing strategies is broadly extended to EndMT-related research25.
To investigate the role of Snail during EndMT, in this study CRISPR/Cas9 gene editing was used to knock out this gene. The data showed that TGF-β2 failed to mediate EndMT in Snail deficient MS-1 cells. This observation demonstrated that Snail is essential for TGF-β2 induced EndMT in MS-1 cells. We used an independent U6-driven sgRNA expression cassette to introduce specific sgRNAs for Cas9 to target Snail. In addition to this method, Ran et al.26 described another strategy for cloning the sgRNA oligos sequence into the Cas9 scaffold to generate a construct containing both Cas9 and gRNAs. Emerging novel approaches allow for CRISPR/Cas to incorporate additional functions. For example, double or triple knockouts can be achieved by delivering more sgRNAs into cells expressing Cas927. The engineered Cas13 protein targets and digests RNA molecules without disrupting endogenous DNA28. Besides knocking out genes with CRISPR/Cas, short hairpin RNAs (shRNAs) can be used as alternatives to stably knock down targeted gene expression29. For all CRISPR/Cas gene editing methods, off-target cleavage should always be taken into consideration. In addition, small interfering RNAs (siRNAs) transiently silence gene expression and the siRNA concentration is diluted with cell division30. Both these methods partially suppress targeted gene expression. In contrast, ectopic gene expression is also used to verify gene function during EndMT/EMT31. This approach can determine if the upregulation of a gene is sufficient to elicit an EndMT response. Therefore, currently there are a multitude of technical strategies that can be used to identify and verify potential regulators of EndMT. Besides, transcriptomic analysis can be a good option in the identification and comprehensive analysis of EndMT related regulators. We recommend using different complementary approaches to investigate the modulation of EndMT.
In summary, we introduced a workflow to identify factors that may play functional roles during TGF-β-induced EndMT. This method can also be used to study whether other stimuli (i.e., cytokines, growth factors, mechanical stimuli, cell-cell interactions) can modulate EndMT, and the interplay of TGF-β with other stimuli. In addition, we highlighted an approach using CRIPSR/Cas gene editing to elucidate whether a certain gene is required for TGF-β-induced EndMT. To illustrate this methodology, we used the strong EndMT inducer TGF-β2 in MS-1 cells, but the protocols can be adapted to other cytokines and other cell types. We expect that this detailed protocol described will serve as a stepping stone for future EndMT-related studies.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The research was supported by CGC.NL and the Netherlands Cardio Vascular Research Initiative: the Dutch Heart Foundation, the Dutch Federation of University Medical Centers, the Netherlands Organization for Health Research and Development, and the Royal Netherlands Academy of Sciences Grant awarded to the Phaedra-Impact (http://www.phaedraresearch.nl). JM is supported by the Chinese Scholarship Council. GSD is supported by a Trampoline grant from AFM-Telethon [22379], FOP Italia and a grant from La Fundació La Marató de TV3 (#202038).
Materials
| 0.45 μm filter | Pall Corporation, USA | 4614 | |
| 10× T4 DNA ligase buffer | Thermofisher Scientific, USA | B69 | |
| 12 mm round glass slice | Knittel Glass,Germany | VD10012Y1A.01 | |
| 20 mL syringe | BD Eclipse, USA | 300629 | |
| 4′,6-diamidino-2-phenylindole (DAPI) | Vector Laboratories, USA | H-1200 | VECTASHIELD Antifade Mounting Media |
| AA19_PLKO vector | Dr. M Gonçalves, Leiden University Medical Center, Netherlands | Gift | |
| Ampicillin | Serva Electrophoresis, USA | 1339903 | |
| Anti-Mouse IgG | GE Healthcare, USA | NA931 | |
| Anti-Rabbit IgG | Cell signaling, USA | 7074 | |
| Agarose | Roche, Switzerland | 11388991001 | |
| Blasticidin | Invitrogen, USA | R21001 | |
| Buffer O (10X) | Thermofisher Scientific, USA | BO5 | |
| BveI (Bspm1) | Thermofisher Scientific, USA | ER 1741 | |
| Confocal microscope | Leica Microsystems, Germany | SP8 | |
| DMEM | Thermo Fisher Scientific, USA | 11965092 | |
| Donkey anti-rat Alexa 488 | Invitrogen, USA | A21208 | |
| FBS | Thermo Fisher Scientific, USA | 16000044 | |
| Formaldehyde | Thermo Fisher Scientific, USA | 28908 | |
| Inverted microscope | Leica Microsystems, Germany | DMi8 | |
| Goat anti-rabbit Alexa 594 | Invitrogen,USA | A11012 | |
| LabNed Plasmid kit | LabNed, USA | LN2400004 | |
| MS-1 cell line | American Type Culture Collection (ATCC) | ||
| Nail polish | HEMA, Netherlands | Transparent | |
| Pecam-1 antibody | Becton Dickinson,USA | 553370 | |
| PEI | Polysciences, USA | 23966-1 | |
| PLV-Cas9 plasmid | Sigma-Aldrich, USA | Cas9BST-1EA | |
| Puromycin | Sigma-Aldrich, USA | P9620 | |
| QIAquick gel extraction kit | Qiagen, Germany | 28706 | |
| Sm22a antibody | Abcam, UK | ab14106 | |
| Snail | Cell signaling, USA | 3879 | |
| T4 DNA ligase | Thermofisher Scientific, USA | EL 0014 | |
| TC20 automated Cell Counter | Bio-Rad, USA | 1450102 | |
| Human TGF-β2 | Joachim Nickel, University of Wurzburg | Gift | Other commercial recommendation: 302-B2, R&D systems |
| Triton X-100 | Merck, USA | 1086031000 | |
| SB431542 | Tocris Bioscience, UK | 1614 | |
| Tris | Roche, Switzerland | 11814273001 |
References
- Ma, J., Sanchez-Duffhues, G., Goumans, M. -. J., Ten Dijke, P. TGF-β-induced endothelial to mesenchymal transition in disease and tissue engineering. Frontiers in Cell and Developmental Biology. 8, (2020).
- Piera-Velazquez, S., Jimenez, S. A. Endothelial to mesenchymal transition: role in physiology and in the pathogenesis of human diseases. Physiological Reviews. 99 (2), 1281-1324 (2019).
- Sánchez-Duffhues, G., de Vinuesa, G. A., ten Dijke, P. Endothelial-to-mesenchymal transition in cardiovascular diseases: developmental signaling pathways gone awry. Developmental Dynamics. 247 (3), 492-508 (2018).
- Evrard, S. M., et al. Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nature Communications. 7 (1), 1-16 (2016).
- Qiao, L., et al. Endothelial fate mapping in mice with pulmonary hypertension. Circulation. 129 (6), 692-703 (2014).
- Zeisberg, E. M., et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nature Medicine. 13 (8), 952-961 (2007).
- Clere, N., Renault, S., Corre, I. Endothelial-to-mesenchymal transition in cancer. Frontiers in Cell and Developmental Biology. 8, (2020).
- Saito, A. EMT and EndMT: regulated in similar ways. The Journal of Biochemistry. 153 (6), 493-495 (2013).
- Bolós, V., et al. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. Journal of Cell Science. 116 (3), 499-511 (2003).
- Aird, W. C. Endothelial cell heterogeneity. Cold Spring Harbor Perspectives in Medicine. 2 (1), 006429 (2012).
- Yang, J., et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nature Reviews Molecular Cell Biology. , 1-12 (2020).
- Doudna, J. A., Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science. 346 (6213), (2014).
- Hsu, P. D., Lander, E. S., Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157 (6), 1262-1278 (2014).
- Arbiser, J. L., et al. Oncogenic H-ras stimulates tumor angiogenesis by two distinct pathways. Proceedings of the National Academy of Sciences USA. 94 (3), 861-866 (1997).
- MA, J., et al. TGF-?-induced endothelial to mesenchymal transition results from a balance between SNAIL and ID factors. Frontiers in Cell and Developmental Biology. 182 (9), (2021).
- Chen, X., et al. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Research. 44 (13), 6482-6492 (2016).
- Hunter, F. W., et al. Functional CRISPR and shRNA screens identify involvement of mitochondrial electron transport in the activation of evofosfamide. Molecular Pharmacology. 95 (6), 638-651 (2019).
- Mao, Y., et al. Lentiviral vectors mediate long-term and high efficiency transgene expression in HEK 293T cells. International Journal of Medical Sciences. 12 (5), 407 (2015).
- Liu, S., et al. Deubiquitinase activity profiling identifies UCHL1 as a candidate oncoprotein that promotes TGFβ-induced breast cancer metastasis. Clinical Cancer Research. 26 (6), 1460-1473 (2020).
- Medici, D., et al. Conversion of vascular endothelial cells into multipotent stem-like cells. Nature Medicine. 16 (12), 1400 (2010).
- Kokudo, T., et al. Snail is required for TGFβ-induced endothelial-mesenchymal transition of embryonic stem cell-derived endothelial cells. Journal of Cell Science. 121 (20), 3317-3324 (2008).
- Dong, J., et al. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biology. 19 (1), 1-20 (2018).
- Oatley, M., et al. Single-cell transcriptomics identifies CD44 as a marker and regulator of endothelial to haematopoietic transition. Nature communications. 11 (1), 1-18 (2020).
- Halaidych, V., et al. Inflammatory responses and barrier function of endothelial cells derived from human induced pluripotent stem cells. Stem Cell Reports. 10 (5), 1642-1656 (2018).
- Li, Y., Lui, K. O., Zhou, B. Reassessing endothelial-to-mesenchymal transition in cardiovascular diseases. Nature Reviews Cardiology. 15 (8), 445-456 (2018).
- Ran, F. A., et al. Genome engineering using the CRISPR-Cas9 system. Nature Protocols. 8 (11), 2281-2308 (2013).
- Grav, L. M., et al. One-step generation of triple knockout CHO cell lines using CRISPR/Cas9 and fluorescent enrichment. Biotechnology Journal. 10 (9), 1446-1456 (2015).
- Abudayyeh, O. O., et al. RNA targeting with CRISPR-Cas13. Nature. 550 (7675), 280-284 (2017).
- Paddison, P. J., et al. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nature Methods. 1 (2), 163-167 (2004).
- Reynolds, A., et al. Rational siRNA design for RNA interference. Nature Biotechnology. 22 (3), 326-330 (2004).
- Lourenço, A. R., et al. C/EBPɑ is crucial determinant of epithelial maintenance by preventing epithelial-to-mesenchymal transition. Nature Communications. 11 (1), 1-18 (2020).

