Automatically Generated
Visualizing and Analyzing the Chemical Space of Natural Product Databases for Drug Discovery
Summary
Here, we provide a methodology that uses different molecular representations to display and analyze the chemical space of natural compound data sets, with a focus on applications related to drug discovery.
Abstract
Chemical space is a multidimensional descriptor space that encloses all possible molecules, and at least 1 x 1060 organic substances with a molecular weight below 500 Da are thought to be potentially relevant for drug discovery. Natural products have been the primary source of the new pharmacological entities marketed during the past forty years and continue to be one of the most productive sources for the creation of innovative medications. Chemoinformatics-based computational tools accelerate the drug development process for natural products. Methods including estimating bioactivities, safety profiles, ADME, and natural product likeness measurement have been used. Here, we go over recent developments in chemoinformatic tools designed to visualize, characterize, and expand the chemical space of natural compound data sets using various molecular representations, create visual representations of such spaces, and investigate structure-property relationships within chemical spaces. With an emphasis on drug discovery applications, we evaluate the open-source databases BIOFACQUIM and PeruNPDB as proof of concept.
Introduction
Natural products (NPs), which are chemical compounds created by living things, have been utilized as traditional treatments for centuries. Individual NPs have been created as medications in the modern era and successfully exploited as lead compounds in drug discovery1. Marine, fungal, bacterial, plant, and endogenous substances created by humans and animals are included in the category of bioactive compounds, as are venoms and poisons produced by various animals2. As a result, for forty years, the number of medications made by NPs represented a significant source of new pharmacological substances3, emphasizing that NPs have been crucial in the development of new medications, particularly for the treatment of cancer and infectious diseases, as well as for other therapeutic conditions like multiple sclerosis and cardiovascular disease4. Furthermore, 64.9% of the 185 small compounds that were authorized to treat cancer between 1981 and 2019 were unmodified NPs or synthetic medicines with an NP pharmacophore3.
Chemoinformatics, a well-established inter-discipline that rests on the concept of chemical space, has been used to analyze and visualize the chemical space of NPs' physicochemical qualities linked to drug-like traits5. Chemoinformatics has shown a substantial impact on drug design and discovery based on NPs6. The chemical space of a group of compounds is not always unique. It will depend on the collection of descriptors used to define it, which means that studying the chemical space of NPs as any other set of compounds, presents particular challenges that rest on molecular representation7. This endeavor can be approached using a variety of molecular descriptors and data visualization techniques. In contrast, the most often utilized techniques are principal component analysis (PCA), scaffold trees, self-organizing maps, generative topographic mapping (GTM), and a novel visualization technique called tree maps (TMAPs)8. Also, the collection, evaluation, and dissemination of NP's chemical information in compound databases is one of the uses of chemoinformatics in NP research. In contrast, with the introduction of big data, this is especially pertinent9.
Here, the open-source NP databases BIOFACQUIM10 and PeruNPDB11 are used to describe the protocol that searches for visualization and characterization of the chemical space of natural compound data sets using various molecular representations, creates visual representations of such spaces and investigates structure-property relationships within chemical spaces, with an emphasis on drug discovery applications.
Protocol
1. Software download and installation
- Make this project's directory fresh. For convenient access, put the executables and files in this directory.
- Install the required software packages after downloading them.
- Download the latest version of The Osiris DataWarrior (OSIRIS) software, which can be found at https://openmolecules.org/datawarrior/
- Download the latest version of The Konstanz Information Miner (KNIME) Analytics Platform, which can be found at https://www.knime.com/
- Download the latest version of The GraphPad Prism software, which can be found at https://www.graphpad.com/
NOTE: The Osiris DataWarrior software and The Konstanz Information Miner (KNIME) Analytics Platform can be used on a personal computer and are free for individual use, while the GraphPad Prism software can be purchased at (https://www.graphpad.com/).
2. Construction and curation of a compound database
NOTE: Find substances and sources that have the necessary data. The user is advised to have the following details for each compound in a spreadsheet.
- Name each compound. Add the names of all the compounds that are described at the source in the first column of a spreadsheet.
- Assign an internal, standardized code if creating an in-house collection, or assign a number that uniquely identifies this compound in the consulted database.
- Provide the structure input using canonical SMILES notation, which can be imported into other molecular editing tools.
- Save the database ideally in .csv format once this data is gathered in the spreadsheet.
- Employ OSIRIS software to generate the dataset's structure data files (SDF), molecular data file (mol), and mol2, which also contain chemical information and are interoperable with most software packages. For this, upload the .csv archive by clicking the File button and then the Open button.
- Upload the dataset to the KNIME analytics platform to improve the data's quality and prevent inaccurate results. For this, upload the .sdf or .mol2 file by clicking the File button and then the Open button.
- Ensure uniformity in chemical structures.
- Examine each chemical structure for valid atom types and valence checks. Standardize the structures by converting them to a canonical tautomeric form, kekulizing aromatic structures, standardizing the positioning of stereo bonds, and turning all implicit hydrogens into explicit hydrogens using the Standardizing Molecular Structures workflow of KNIME.
- Find and eliminate duplicates after the molecules have been correctly standardized by employing the Standardizing Molecular Structures workflow of KNIME. Utilize InChI keys as a linear notation to locate various protonation states and tautomers.
- Eliminate duplicates.
- Enumerate tautomers and stereoisomers. This step is crucial in virtual screening studies, especially when using search methods such as docking or pharmacophore-based filtering.
3. Molecular descriptors and diversity analysis
NOTE: Molecular descriptors, such as physicochemical qualities, and molecular fingerprints and chemical scaffolds, are the most common approaches to represent molecules in chemoinformatic applications. Analysis can be performed here: http://132.248.103.152:3838/PUMA/. All steps described below are detailed on the PUMA website.
- Calculate the six most prevalent physicochemical qualities of pharmacological relevance: molecular weight (MW), octanol/water partition coefficient (clogP), topological surface area (TPSA), aqueous solubility (clogS), number of H-bond donor atoms (HBD), and number of H-bond acceptor atoms (HBA). Refer to the PUMA website for more information.
- Calculate the 166-bit MACCS keys, the pairwise Tanimoto similarity, and extended connectivity fingerprints of diameter 4 (ECFP4), along with other circular fingerprints suitable for virtual screening, activity landscape modeling, and structure-activity relationships (SAR) research.
- Compute a central tendency statistic for each pairwise comparison. Ensure diversity in the dataset with a smaller mean or median contrary to Euclidean distance or any general distance metric.
- Check if the calculated values have been recorded in the literature or computed for other reference databases for comparison purposes. For this, consult websites such as PubChem or CHEMBL.
- Generate violin plots for visualization within the GraphPad Prism software, displaying the maximum and minimum values.
4. Visualization of the chemical space
NOTE: It is possible to condense the majority of the pertinent data into a small number of variables using PCA and other dimensionality reduction techniques. Visualizations of the chemical space are therefore made possible.
- Select all the six descriptors to determine similarity or distance. Create the similarity (or distance) matrix accordingly.
- Perform PCA analysis on the matrix. Select two or three main components for plotting. Consider the proportion of variance captured by each primary component.
- Generate two or three-dimensional scatter-plot representations for PCA using the Plotly KNIME node.
5. Consensus diversity plots
NOTE: Visual representations have been developed to summarize a few characteristics that can be used to quantify variety. The consensus diversity plots (CDPs)12 analysis can be performed here http://132.248.103.152:3838/CDPlots/.
- Create a plot with the number of compounds in the database to determine the data point size. Use the diversity of molecular fingerprints for the x-axis, diversity of scaffolds for the y-axis, diversity based on physicochemical properties for the color continuous scale, and the relative number of compounds in the dataset for data point size.
- Generate the multiple-variable plot using the GraphPad Prism software.
Representative Results
Molecular properties and visualization of the chemical space
All compounds in the BIOFACQUIM10, PeruNPDB11, and FDA13 datasets had six physicochemical properties calculated for them. These qualities were then plotted onto violin plots, which allow one to see how the properties of the three studied datasets are distributed (Figure 1). The distribution profiles of the six physicochemical parameters of pharmaceutical interest, namely molecular weight (MW), octanol/water partition coefficient (clogP), topological surface area (TPSA), aqueous solubility (clogS), number of H-bond donor atoms (HBD), and number of H-bond acceptor atoms (HBA), differ between the datasets. However, the TPSA results demonstrated significant variances when comparing the BIOFACQUIM and FDA datasets to PeruNPDB. Using PCA, the dataset's chemical space visualization was carried out. However, 3D-visual PCA analysis reveals that molecules in both datasets of NPs roughly overlap chemical space with the FDA's collection of approved pharmaceuticals. While in some areas, chemicals from PeruNPDB or BIOFACQUIM predominate (Figure 2).
Diversity analysis
In addition, a CDP based on molecular fingerprints, scaffolds, and physicochemical attributes was utilized to assess the diversity of the datasets. The property-based diversity of the databases from PeruNPDB, BIOFAQUIM, and FDA was calculated using the Euclidean distance of the scaled properties. Besides, a CDP based on molecular fingerprints, scaffolds, and physicochemical attributes was utilized to assess the diversity of the datasets. The property-based diversity of the databases from PeruNPDB, BIOFAQUIM, and FDA was calculated using the Euclidean distance of the scaled properties. The values on the color CD plot are represented by data points on a continuous color scale. Brighter hues denote greater diversity, whereas darker colors denote less diversity. Last but not least, various point sizes are utilized to show the relative number of compounds in each database, with smaller data points representing databases with fewer molecules. Since it was discovered in the region where the greatest diversity in scaffold and fingerprints should be located, the findings showed that the compounds in the PeruNPDB had the greatest global diversity (Figure 3).
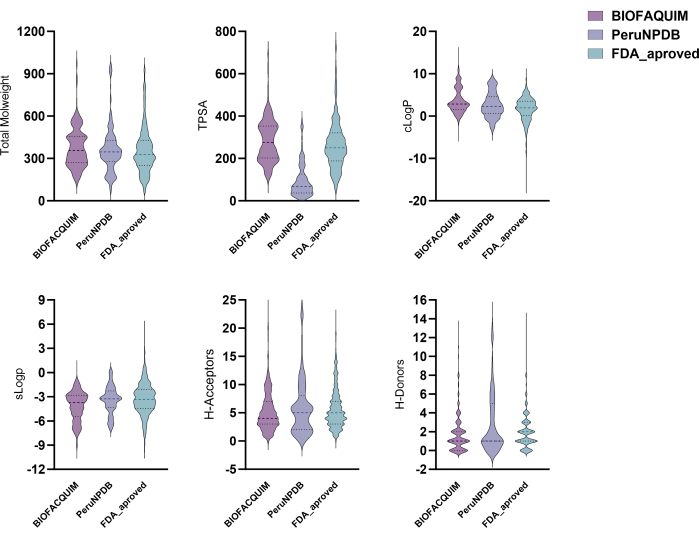
Figure 1: Violin plots for the physicochemical properties. Violin plots for the physicochemical properties of BIOFACQUIM, PeruNPDB, and FDA datasets. Please click here to view a larger version of this figure.
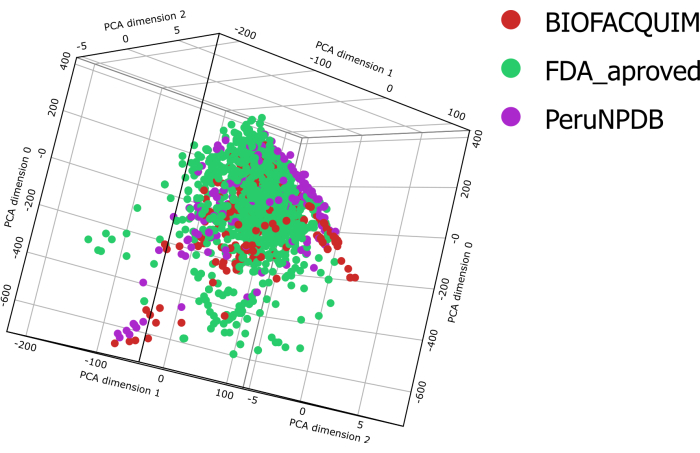
Figure 2: Visual representation of the chemical space. Visual representation of BIOFACQUIM, PeruNPDB, and FDA datasets based on principal components of six properties of pharmaceutical relevance. Please click here to view a larger version of this figure.
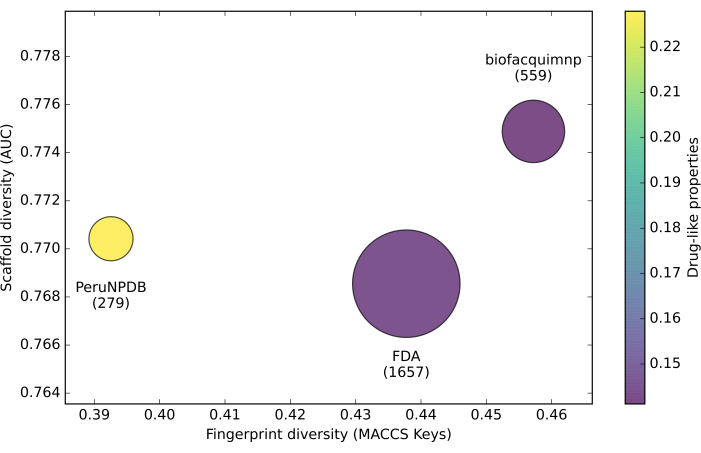
Figure 3: Consensus diversity plot. Consensus diversity plot comparing the global diversity of BIOFACQUIM, PeruNPDB, and FDA datasets. Please click here to view a larger version of this figure.
Discussion
Due to its many potential uses, such as compound classification, compound selection, exploring structure-activity links, and navigating through structure-property interactions, the concept of chemical space is nowadays widely employed in the drug discovery and development process14. Also, the creation of NP databases is a fundamental procedure to perform various computational studies, including the design of chemical libraries, characterization and comparison of the chemical space, the study of SAR, and virtual screening, among other studies, as a result of the increase in the amount of chemical information. In contrast, training in artificial intelligence (AI) algorithms is yet another crucial application. AI refers to a group of computational techniques that enable machines to mimic human cognitive processes, including problem-solving and learning from experience15,16.
Though chemical compounds databases, including NP databases, are important tools in drug discovery, it is also feasible to detect potential hit molecules using a variety of virtual screening techniques17. Additionally, several drug candidates have been found in NP databases that have the potential to treat illnesses, including coronavirus disease18, Alzheimer's disease19, and leishmaniasis20, among others. However, due to present constraints in processing "big data", the chemical space of all potential molecules in a particular biological or environmental sample might be extremely large and mostly unexplored21. Although there are no unique or universal techniques for chemical space representations, one widely used way involves creating similarity matrices that include all pairwise comparisons22. The majority of the pertinent information can be reduced into a small number of variables (albeit losing information) using PCA and other dimensionality reduction techniques, enabling the visualization of the chemical space23.
A chemical library's diversity can be evaluated in a variety of ways, largely depending on the data being examined and, most importantly, the goal of the study. Molecular representation is a crucial component of diversity analysis in addition to the diversity measure24. Although chemical scaffolds and molecular descriptors are the two approaches used to represent molecules most frequently in chemoinformatic analysis, some of them have the drawback of being more challenging to comprehend25 and do not necessarily identify the collections; for instance, it is typical for various compounds to have extremely comparable property profiles. Therefore, considering various structure representations offers a more comprehensive picture of the diversity of compound libraries. This is the basis of the concept of chemical multiverse, whichan be defined as a group or collection of chemical spaces for the same data set, each one defined by a set of descriptors26.
Because CDPs use multiple representations that can be divided into three or two dimensions to analyze the global diversity of compound data sets using a variety of metrics, they help compare and categorize chemical libraries12.
Disclosures
The authors have nothing to disclose.
Acknowledgements
HLBC and MACH thank the funding of Universidad Catolica de Santa Maria (grants 27499-R-2020, 27574-R-2020, 7309-CU-2020, and 28048-R-2021). JLMF thanks the funding of DGAPA, UNAM, Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT), grant No. IN201321.
Materials
| GraphPad Prism | GraphPad Prism | https://www.graphpad.com/ | |
| KNIME platform | KNIME | https://www.knime.com | |
| Osiris DataWarrior (OSIRIS) software | openmolecules.org | https://openmolecules.org/datawarrior/ | |
| PUMA | PUMA: Platform for Unified Molecular Analysis | http://132.248.103.152:3838/PUMA/ |
References
- Boufridi, A., Quinn, R. J. Harnessing the properties of natural products. Annu Rev Pharmacol Toxicol. 58, 451-470 (2018).
- Gómez-García, A., et al. Navigating the chemical space and chemical multiverse of a unified Latin American natural product database: LANaPDB. ChemRxiv. , (2023).
- Newman, D. J., Cragg, G. M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 83 (3), 770-803 (2020).
- Atanasov, A. G., Zotchev, S. B., Dirsch, V. M., Supuran, C. T. Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov. 20 (3), 200-216 (2021).
- Medina-Franco, J. L., Saldívar-González, F. I. Cheminformatics to characterize pharmacologically active natural products. Biomolecules. 10 (11), 1566 (2020).
- Chen, Y., Garcia De Lomana, M., Friedrich, N. O., Kirchmair, J. Characterization of the Chemical Space of Known and Readily Obtainable Natural Products. J Chem Inf Model. 58 (8), 1518-1532 (2018).
- Gaytán-Hernández, D., Chávez-Hernández, A. L., López-López, E., Miranda-Salas, J., Saldívar-González, F. I., Medina-Franco, J. L. Art driven by visual representations of chemical space. ChemRxiv. , (2023).
- Zabolotna, Y., Ertl, P., Horvath, D., Bonachera, F., Marcou, G., Varnek, A. NP Navigator: A new look at the natural product chemical space. Mol Inform. 40 (9), e2100068 (2021).
- Martinez-Mayorga, K., Madariaga-Mazon, A., Medina-Franco, J. L., Maggiora, G. The impact of chemoinformatics on drug discovery in the pharmaceutical industry. Expert Opin Drug Discov. 15 (3), 293-306 (2020).
- Pilón-Jiménez, B., Saldívar-González, F., Díaz-Eufracio, B., Medina-Franco, J. BIOFACQUIM: A Mexican compound database of natural products. Biomolecules. 9 (1), 31 (2019).
- Barazorda-Ccahuana, H. L., et al. PeruNPDB: the Peruvian natural products database for in silico drug screening. Sci Rep. 13 (1), 7577 (2023).
- González-Medina, M., Prieto-Martínez, F. D., Owen, J. R., Medina-Franco, J. L. Consensus diversity plots: a global diversity analysis of chemical libraries. J Cheminform. 8, 63 (2016).
- Irwin, J. J., et al. ZINC20-A free ultralarge-scale chemical database for ligand discovery. J Chem Inf Model. 60 (12), 6065-6073 (2020).
- Naveja, J. J., Medina-Franco, J. L. Finding constellations in chemical space through core analysis. Front Chem. 7, 510 (2019).
- Cavasotto, C. N., Di Filippo, J. I. Artificial intelligence in the early stages of drug discovery. Arch Biochem Biophys. 698, 108730 (2021).
- Rosén, J., Gottfries, J., Muresan, S., Backlund, A., Oprea, T. I. Novel chemical space exploration via natural products. J Med Chem. 52 (7), 1953-1962 (2009).
- Sliwoski, G., Kothiwale, S., Meiler, J., Lowe Jr, E. W. Computational methods in drug discovery. Pharmacol Rev. 66 (1), 334-395 (2014).
- Goyzueta-Mamani, L. D., Barazorda-Ccahuana, H. L., Mena-Ulecia, K., Chávez-Fumagalli, M. A. Antiviral activity of metabolites from Peruvian plants against SARS-CoV-2: An in silico approach. Molecules. 26 (13), 3882 (2021).
- Goyzueta-Mamani, L. D., et al. In silico analysis of metabolites from Peruvian native plants as potential therapeutics against Alzheimer’s disease. Molecules. 27 (3), 918 (2022).
- Barazorda-Ccahuana, H. L., et al. Computer-aided drug design approaches applied to screen natural product’s structural analogs targeting arginase in Leishmania spp. F1000Research. 12, 93 (2023).
- McGrady, M. Y., Colby, S. M., Nuñez, J. R., Renslow, R. S., Metz, T. O. AI for chemical space gap filling and novel compound generation. arXiv. , (2022).
- Medina-Franco, J., Martinez-Mayorga, K., Giulianotti, M., Houghten, R., Pinilla, C. Visualization of the chemical space in drug discovery. Curr Comput Aided-Drug Des. 4 (4), 322-333 (2008).
- Osolodkin, D. I., Radchenko, E. V., Orlov, A. A., Voronkov, A. E., Palyulin, V. A., Zefirov, N. S. Progress in visual representations of chemical space. Expert Opin Drug Discov. 10 (9), 959-973 (2015).
- Sheridan, R. P., Kearsley, S. K. Why do we need so many chemical similarity search methods. Drug Discov Today. 7 (17), 903-911 (2002).
- Singh, N., Guha, R., Giulianotti, M. A., Pinilla, C., Houghten, R. A., Medina-Franco, J. L. Chemoinformatic analysis of combinatorial libraries, drugs, natural products, and molecular libraries Small Molecule Repository. J Chem Inf Model. 49 (4), 1010-1024 (2009).
- Medina-Franco, J. L., Chávez-Hernández, A. L., López-López, E., Saldívar-González, F. I. Chemical multiverse: An expanded view of chemical space. Mol Inform. 41 (11), e2200116 (2022).

.
