Automatically Generated
Isolation and Culture of Three Kinds of Umbilical Cord Mesenchymal Stem Cells
Summary
The present protocol describes the isolation and culture of mesenchymal stem cells from the umbilical cord arteries, vein, and Wharton’s jelly.
Abstract
Umbilical cord mesenchymal stem cells (UC-MSCs) are an important cell source for regenerative medicine. UC-MSCs can be isolated from the umbilical cord Wharton’s jelly, as well as from the umbilical arteries and umbilical vein. They are known as perivascular stem cells obtained from umbilical arteries (UCA-PSCs), perivascular stem cells obtained from the umbilical vein (UCV-PSCs), and mesenchymal stem cells obtained from Wharton’s jelly (WJ-MSCs). UCA-PSCs and UCV-PSCs are pericytes derived from perivascular regions that are progenitors of MSCs. Isolation and culture of the three kinds of cells is an important source for studying stem cell transplantation and repair. The present protocol focuses on the isolation and culture of cells through mechanical separation, adherent culture, and cell crawling out. Through this technique, the three different types of stem cells can be derived. Cell surface markers were detected by flow cytometry. The stem cells were detected for multilineage differentiation potential by adipogenic, osteogenic, and neural-like differentiation, which is consistent with the phenotype of MSCs. This experimental protocol expands the source of UC-MSCs. In addition, the cell isolation method provides a basis for further study of regenerative medicine and other applications.
Introduction
Human umbilical cord mesenchymal stem cells (UC-MSCs) are widely used in regenerative medicine because of their noninvasive operation, low immunogenicity, and lack of ethical dispute1. In many studies, UC-MSCs isolated from Wharton’s jelly (WJ) can attach to the wall, undergo multi-differentiation, and express markers of mesenchymal stem cells (MSCs)2. However, almost all MSCs originate from the perivascular region3. Pericytes, as a subset of perivascular cells, are progenitor cells of MSCs4. Therefore, UC-MSCs can be isolated from the umbilical cord WJ, umbilical arteries (UCAs), and umbilical vein (UCV), known as UCA-PSCs, UCV-PSCs, and WJ-MSCs, respectively5. This method aimed to isolate and culture the three different types of stem cells. The isolation and culture of UCA-PSCs, UCV-PSCs, and WJ-MSCs are very important to provide more sources of MSCs.
The present study describes the isolation, culture, and future application of UCA-PSCs, UCV-PSCs, and WJ-MSCs, which have cellular adhesion, express the markers of MSCs, and have multidirectional differentiation. The isolated stem cells were observed under microscopy and subjected to cell culture, cell passage, cell cryopreservation, and cell recovery. The rationale behind the use of this technique was cells crawling out from tissue. Compared to the previous method, such as flow cytometry or immunomagnetic bead techniques, which were complex and expensive6, the US-MSCs can be massively isolated by the adherent separation and cell crawling method; these were used in the previous study5. Flow cytometry analysis was performed on the derived stem cells to detect whether these cells express MSC markers. Multidirectional differentiation of the stem cells was introduced to detect whether the three kinds of cells have the potential to differentiate into adipocytes, osteoblasts, and neuroblasts. The isolation and culture of three types of stem cells from the umbilical cord were important in clinical use and helpful for researchers for diverse future applications.
Protocol
All experimental procedures were approved by the Clinical Research Ethics Committee, Third Affiliated Hospital, Soochow University. Informed written consent was obtained from the human subjects. Individuals with full-term vaginal delivery or cesarean section were included in the present study to obtain the umbilical cord. The umbilical cord comes from healthy newborns without gender bias. The neonate had an Apgar score of 8-10. The Apgar score is a quick test for newborns given soon after their birth. This test checks a baby's muscle tone, heart rate, and other signs to see if any additional medical or emergency care is needed7. On the other hand, patients with major diseases, such as heart, liver, kidney, or other infectious diseases, were excluded from the present study.
1. Collection of human umbilical cord
- Place the newborn's umbilical cord into phosphate buffered saline (PBS) and transport at 4 °C.
NOTE: Three umbilical cords from three donors were used in the present study. - Remove the umbilical cord blood immediately under sterile conditions, and cut along the long axis of the blood vessels in 20 cm of the umbilical cord. Separate the appropriate 20 cm of the umbilical arteries and vein, and remove the Wharton's jelly on the surface carefully.
NOTE: Explant cultures were obtained from each region, minced into small pieces (1-2 mm3) to avoid cross contamination among the three types of stem cells. - Cut the umbilical arteries, vein, and Wharton's jelly to a size of 1-2 mm3 and inoculate them into 100 mm cell culture dishes. Perform the inoculation of the umbilical arteries, vein, and Wharton's jelly using tweezers following a previously published report5. Do not precoat the 100 mm culture plates.
NOTE: The interval between the umbilical arteries, vein, or Wharton's jelly was about 1 cm. A too long collection time before separating cells may affect the cell viability and amount. It is recommended to separate the arteries and vein from the umbilical cord immediately. The collection time was up to 4 h.
2. Isolating and culturing of UCA-PSCs, UCV-PSCs, and WJ-MSCs
- Place the cell culture dishes upside down for 3 h in a 5% CO2 incubator at 37 °C to ensure the umbilical arteries, vein, and Wharton's jelly tissue attach to the cell culture dishes tightly before the culture medium is added.
- After 3 h, return the cell culture dishes to normal placement. Add 5 mL of cell culture medium (LG-DMEM, 10% FBS, 100 IU/mL of penicillin, and 100 µg/mL of streptomycin, see Table of Materials) to the cell culture dish.
- Add 3 mL and 2 mL of the culture medium into the cell culture dish on the 3rd and 7th day of the first week, respectively. Be careful not to touch the tissue blocks of umbilical cord arteries, vein, and Wharton's jelly.
- Change half of the liquid medium twice in the second week and the entire liquid medium twice in the third week.
- Observe the cells crawling out of the tissue blocks at approximately 7 days post-culture, and then discard the tissue blocks when the cells are 80% fused at ~2 weeks.
- When the cells are 100% fused, passage them at a ratio of 1:3 with 2 mL of 0.05% trypsin after centrifugation at 600 x g for 4 min at room temperature.
NOTE: The primary cells isolated from an umbilical cord were about 2 x 106 UCA-PSCs, 1 x 106 UCV-PSCs, and 2 x 106 WJ-MSCs. The 2 x 106 UCA-PSCs, 1 x 106 UCV-PSCs, and 2 x 106 WJ-MSCs were isolated from 20 cm of umbilical cord. The cells crawled out in all three umbilical cords, which indicated results between isolations were repeatable. - Seed the cells, maintaining a concentration of 5 x 105/100 cm2 in the culture dish. Preserve the 5 x 105 cells in 1 mL of cryopreservation solution (90% FBS, 10% DMSO). Third- to fifth-passage cells were used in the present study.
- When the cells are 100% fused, passage them at a ratio of 1:3 with 2 mL of 0.05% trypsin after centrifugation at 600 x g for 4 min at room temperature.
3. Detection of MSC surface markers by flow cytometry
- Digest the third-generation cells with 0.05% trypsin to obtain a single-cell suspension. Centrifuge the cells at 600 x g for 4 min at room temperature and discard the cell supernatant using a Pasteur pipette. Then resuspend 1 x 105 cells in 100 µL of PBS (1% FBS). Asses the cell number by counting the cells using a hemocytometer following a previously published report8.
- Incubate the 1 x 105 cells with various phycoerythrin antibodies against CD13 (FITC, 0.1 mg/mL), CD34 (FITC, 0.1 mg/mL), CD45 (FITC, 0.1 mg/mL), CD73 (FITC, 0.1 mg/mL), and HLA-DR (FITC, 0.1 mg/mL) in the dark for 30 min. For controls, incubate 1 x 105 cells with FITC-IgG (FITC Mouse Anti-Human IgG, 0.1 mg/mL) (see Table of Materials).
- Centrifuge the cells at 600 x g for 4 min at room temperature, and then discard the cell supernatant using a Pasteur pipette. Resuspend 1 x 105 cells in 500 µL of PBS (1% FBS), detect by flow cytometry, and analyze by FlowJo2 (see Table of Materials).
4. Differentiation of UCA-PSCs, UCV-PSCs, and WJ-MSCs into adipocytes
- Inoculate the third-generation cells into 24-well plates at a density of 6 x 104 cells/cm2 with 500 µL of cell medium (LG-DMEM, 10% FBS, 100 IU/mL penicillin, and 100 µg/mL streptomycin, see Table of Materials) per well in a 5% CO2 incubator at 37 °C.
- Change the cell medium after 24 h, and change the control group to cell medium (LG-DMEM, 10% FBS, 100 IU/mL penicillin, and 100 µg/mL streptomycin). Conduct the experiments after the cells are 80% fused in 24-well plates.
NOTE: The cells in the groups that induced adipogenesis were changed to an adipogenic induction medium (adipogenic differentiation kit, see Table of Materials). The medium of the cells in both groups was changed every 3 days. - Observe lipid droplets in cells at approximately 21 days post-induction in the adipogenic induction group and then terminate the adipogenic induction.
NOTE: The adipogenic medium was replaced by cell culture medium to terminate the adipogenic induction2,5. In addition, the cells in the control group showed no significant change. - Wash the cells three times with PBS for 5 min and then fix with 4% paraformaldehyde for 15 min at room temperature. Then, wash the cells three times with PBS for 5 min each.
- Discard the PBS and dry the cells at room temperature for approximately 20 min. Then, add 500 µL of oil red O solution (0.5%) (see Table of Materials) to the well for 10 min.
- Remove the oil red O solution and wash the cells with PBS three times for 5 min each. Observe the cells under a microscope.
5. Differentiation of the stem cells into osteoblasts
- Place 500 µL of 0.1% gelatin (see Table of Materials) per well in 24-well culture plates and discard it after 30 min.
- Inoculate 6 x 104/cm2 of third-generation cells into 24-well culture plates, and add 500 µL cell culture medium or osteogenic induction medium (Osteogenic Differentiation Kit, see Table of Materials) to each well in a 5% CO2 incubator at 37 °C. In the control group, change the cell culture medium every 3 days, while also replacing the osteogenic induction medium in the osteogenic differentiation group every 3 days.
- After 3-4 weeks, check for black calcium deposition in the cells. Wash the well with PBS three times for 5 min each.
- Fix the cells at room temperature with 4% paraformaldehyde for 15 min and wash with PBS three times for 5 min each. Add alizarin red staining solution (1%) to the well for 20 min at room temperature, and then discard it.
- After drying for 30 min at room temperature, observe the cells under a microscope.
6. Differentiation of the stem cells into neurons
- Inoculate the 6 x 104/cm2 third-generation cells into 24-well culture plates, where sterile cover slides treated with L-polylysine were preplaced. Place the cell climbing sheets (see Table of Materials) in the 24-well plates to differentiate the cells into neurons. Culture the cells on the cell climbing sheets.
- Then, add 500 µL of cell culture medium to the well, and replace the cell culture medium with 500 µL of neurogenic induction medium9 (see Table of Materials) after 24 h.
- After induction for 24 h, add 10-7 mol/L of ATRA and 10 ng/mL of bFGF (see Table of Materials) to the solution to maintain the induced differentiation of neurons.
- Terminate the induction when obvious cellular synapses are formed. Check for the obvious cellular synapses using microscopy technique (see Table of Materials).
NOTE: Immunofluorescence staining was used to identify neuron-specific markers of neuron-specific enolase (NSE, see Table of Materials), which is a marker of neurons2. - Perform confocal imaging on the cell climbing sheets.
7. Immunofluorescence staining
- After discarding the cell culture solution, wash the cells with 500 µL of PBS three times for 5 min each.
- Then, add 500 µL of 4% paraformaldehyde to the well, and fix the cells at room temperature for 45 min.
- Wash the cells with PBS three times for 5 min each.
- Permeabilize the cells with 100 µL of 0.5% Triton-X at room temperature for 5 min.
- Wash the cells with 500 µL PBS with 1% BSA three times for 5 min each.
- Incubate the cells with 500 µL of PBS with 3% BSA and 0.1% Triton X-100 per well at 37 °C for 45 min to block the nonspecific binding site.
- Dilute the primary antibody against NSE (1:100, see Table of Materials) with PBS (1:100) containing 1% BSA and 0.1% Triton X-100, and incubate the cells with the primary antibody overnight at 4 °C.
- After discarding the primary antibody, wash the cells with 500 µL of PBS containing 1% BSA three times for 5 min each.
- Dilute the fluorescent secondary antibody (goat Anti-Mouse IgG H&L, Alexa Fluor 488, see Tabel of Materials) with PBS (1:200) containing 1% BSA and 0.1% Triton X-100 at 1:200, and incubate the cells with the secondary antibody at 37 °C in the dark for 60 min.
- Wash the cells with 500 µL of PBST (PBS containing 0.1% Tween-20) per well three times in the dark for 5 min each.
- Then, add 200 µL of DAPI staining (PBST diluted, 0.1 µg/mL) to the well to stain the nucleus at room temperature without light for 5 min.
- Wash the cells with 500 µL of PBST three times in the dark for 5 min each.
- Add the anti-fluorescence quenching agent to the slide and then gently cover the slide with a cover glass. Add nail polish drops around the cover glass to fix it.
- Observe the glass with a laser confocal microscope. Use the following microscopy settings: green channel with 488 nm laser and DAPI channel with 405 nm.
8. Proliferation assay
- Seed 2 x 103 cells per well into 96-well plates. Add 100 µL of cell culture medium into each well.
- Add 10 µL of Cell-counting kit-8 (CCK-8, see Table of Materials) into each well.
- After 2 days of incubation, measure the OD value at 450 nm on a microplate reader on days 1-7.
- Analyze the OD values of the different types of stem cells using statistical software (see Table of Materials) and present them as means ± SD. Perform one-way analysis of variance (ANOVA). Consider P value < 0.05 to be statistically significant.
Representative Results
Isolation and culture of UCA-PSCs, UCV-PSCs, and WJ-MSCs from the umbilical cord
The umbilical arteries, umbilical vein, and Wharton's jelly were mechanically separated from the umbilical cord and cut into 2-3 cm3 pieces. The distance between arteries, vein, or Wharton's jelly tissue blocks was approximately 1 cm, arranged in a quincunx shape (Figure 1A–C). The three kinds of stem cells were isolated by a tissue-attached culture method5. After approximately 1 week of attached culture, the cells were observed to crawl out radially, exhibiting a long spindle shape and rapid proliferation (Figure 1D–F). The cells reached 70%-80% confluence at approximately 2 weeks, and the growth rate was accelerated after passage. The cells either grew in an even spindle, parallel arrangement, or vortex growth (Figure 1G–I). In addition, the different stem cells had similar proliferation tendencies as demonstrated by the CCK-8 assay (Supplementary Figure 1). However, on days 4 and 5, UCA-PSCs had a significant higher growth rate compared to UCV-PSCs (P < 0.05).
UCA-PSCs, UCV-PSCs, and WJ-MSCs expressed MSC surface markers
The surface markers of third-generation stem cells were identified by flow cytometry. As shown in Figure 2A–O, UCA-PSCs, UCV-PSCs, and WJ-MSCs all highly expressed the MSC-specific surface markers CD13 and CD73, but did not express CD34 and CD45, which are endothelial cell markers and hematopoietic stem cell markers. The cells did not express HLA-DR, a hematopoietic stem cell marker2. The characterization of Adipose-derived stem cells (ADSCs) as a control was displayed in Supplementary Figure 2A-E, which indicates that the stem cells expressed the surface marker of MSCs consistent with that of ADSCs. MSCs were incubated with FITC-IgG and were negative for the FITC signal (Supplementary Figure 3A). In addition, the results of three repeated experiments were analyzed in Supplementary Figure 4, which proved the method was reproducible.
UCA-PSCs, UCV-PSCs, and WJ-MSCs possessed adipogenic differentiation potential
After adipogenic induction, the morphology of the isolated stem cells changed from long spindle to round. After approximately 12 days of adipogenic induction, round lipid droplets were observed in the cells, which had strong refraction. Some lipid droplets fused to form large lipid droplets. On approximately the 14th day, the three kinds of cells showed red lipid droplets of different sizes by oil red O staining (Figure 3A-C). The characterization of ADSCs as a control was displayed in Supplementary Figure 2F, which indicates that the different stem cells possessed adipogenic differentiation potential consistent with that of ADSCs. For adipogenic differentiation, there were no lipid droplets in the human endometrial stromal cells (ESCs) (Supplementary Figure 3B).
UCA-PSCs, UCV-PSCs, and WJ-MSCs have osteogenic differentiation potential
After osteogenic induction, the cell morphology changed from a long spindle to a lamellar structure, and the extracellular matrix began to deposit. After osteogenic induction for 10 days, calcium nodules appeared in the three kinds of cells. At approximately 21 days after induction, the cell morphology was polygonal, and a calcium nodule-like structure was seen in the center of the cells. The cells showed calcified nodules in the middle, as shown by Alizarin red staining (Figure 4A-C). The characterization of ADSCs as a control was displayed in Supplementary Figure 2G, which indicates that stem cells possessed osteogenic differentiation potential consistent with that of ADSCs. For osteogenic differentiation potential, there were no calcified nodules in the ESCs (Supplementary Figure 3C).
UCA-PSCs, UCV-PSCs, and WJ-MSCs have neurogenic differentiation potential
The three kinds of stem cells were induced by neurogenic induction solution; the spindle-shaped fibroblast-like cells became floated ball-shaped neurosphere-like cells. The cellular synapses were formed in all the cell variants. The morphology of the cells gradually became stellate and interconnected, and had nerve-like characteristics and enhanced refraction. The results of cellular immunofluorescence showed that the stem cells expressed neuron-specific markers of NSE after induction (Figure 5A-C). The results showed that the cells had multidirectional differentiation potential after induction. The characterization of ADSCs as a control was displayed in Supplementary Figure 2H, which indicates that the stem cells possessed neurogenic differentiation potential consistent with that of ADSCs. For neurogenic differentiation potential, there was no NSE signal in the ESCs (Supplementary Figure 3D).
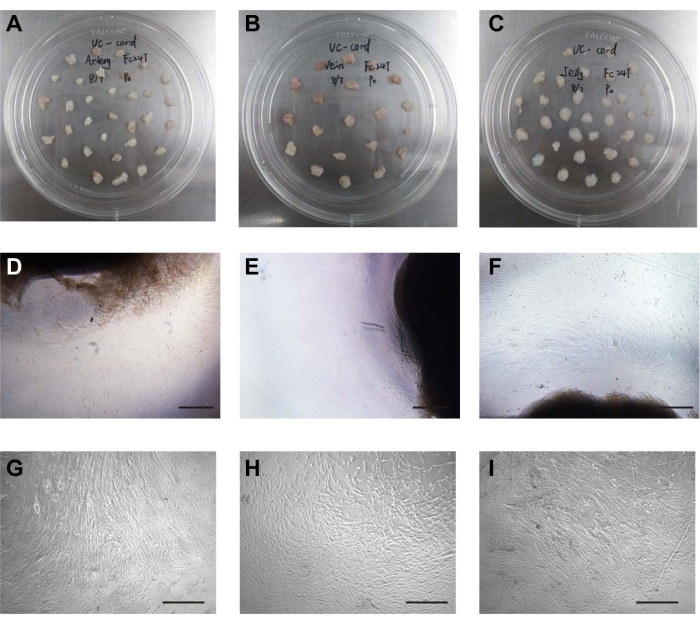
Figure 1: Isolation and culture of cells. (A) The umbilical arteries were placed in the cell culture dish. (B) The umbilical vein was placed in the cell culture dish. (C) Wharton's jelly was placed in the cell culture dish. (D) The stem cells crawled out of the umbilical artery tissue. Scale bar = 500 µm. (E) The stem cells crawled out of umbilical vein tissue. Scale bar = 500 µm. (F) The stem cells crawled out of the Wharton's jelly tissue. Scale bar = 500 µm. (G) Morphology of cultured UCA stem cells after passage. Scale bar = 500 µm. (H) Morphology of cultured UCV stem cells after passage. Scale bar = 500 µm. (I) Morphology of cultured WJ stem cells after passage. Scale bar = 500 µm. Please click here to view a larger version of this figure.
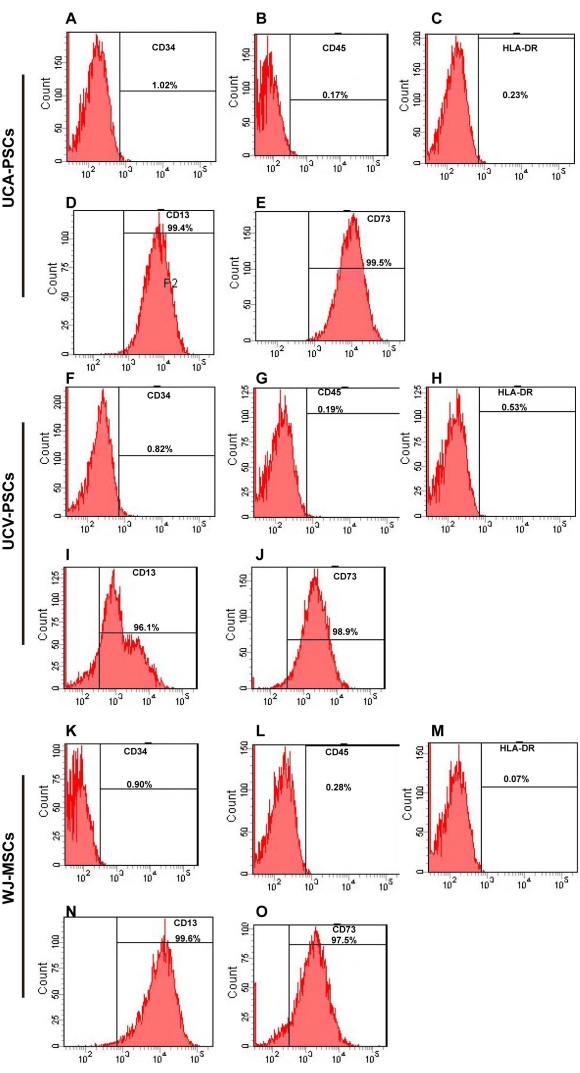
Figure 2: Flow cytometry analysis of UCA-PSC, UCV-PSC, and WJ-MSC surface markers. (A-E) UCA-PSCs were negative for CD34, CD45, and HLA-DR. UCA-PSCs were positive for CD13 and CD73. (F–J) UCV-PSCs were negative for CD34, CD45, and HLA-DR. UCV-PSCs were positive for CD13 and CD73. (K–O) WJ-MSCs were negative for CD34, CD45, and HLA-DR. WJ-MSCs were positive for CD13 and CD73. The x-axis is the amount of fluorescence. Please click here to view a larger version of this figure.
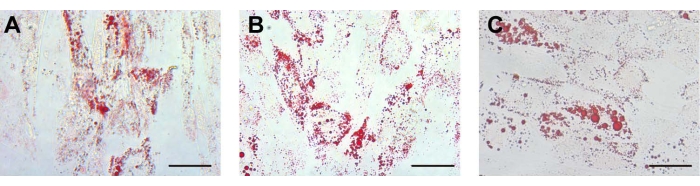
Figure 3: Adipogenic differentiation. For adipogenic differentiation, the lipid droplets in the (A) UCA-PSCs, (B) UCV-PSCs, and (C) WJ-MSCs cultured in an adipogenic induction medium were stained with oil red O. Scale bar, 50 µm. Please click here to view a larger version of this figure.
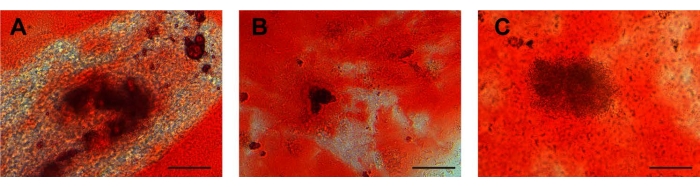
Figure 4: Osteogenic differentiation. Calcium deposits in (A) UCA-PSCs, (B) UCV-PSCs, and (C) WJ-MSCs cultured in an osteogenic induction medium were stained with Alizarin red for osteogenic differentiation. Purple-red clumps are calcium nodules. Scale bar = 50 µm. Please click here to view a larger version of this figure.
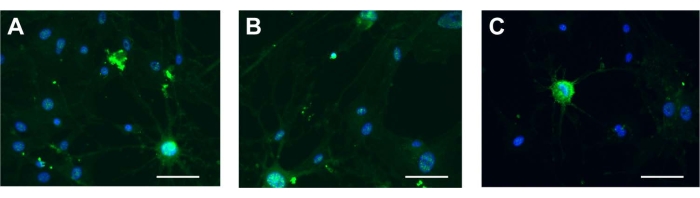
Figure 5: Neurogenic differentiation. NSE in (A) UCA-PSCs, (B) UCV-PSCs, and (C) WJ-MSCs cultured in a neurogenic induction medium. Green shows NSE stain, and blue shows nucleus. Scale bar = 50 µm. Please click here to view a larger version of this figure.
Supplementary Figure 1: Cell proliferation. CCK-8 assays reflected that the different stem cells had similar proliferation tendencies. On days 4 and 5, UCA-PSCs had a significant higher growth rate than UCV-PSCs. The error bars represent the means ± SE of the three independent experiments performed in triplicate. P < 0.05. Please click here to download this File.
Supplementary Figure 2: Characterization of Adipose-derived stem cells (ADSCs) as the positive control. (A–E) ADSCs were negative for CD34, CD45, and HLA-DR. ADSCs were positive for CD13 and CD73. (F) For adipogenic differentiation, the lipid droplets in ADSCs cultured in an adipogenic induction medium were stained with oil red O. Scale bar = 50 µm. (G) Calcium deposits in ADSCs cultured in an osteogenic induction medium were stained with Alizarin red for osteogenic differentiation. Scale bar = 50 µm. (H) NSE in ADSCs cultured in neurogenic induction medium. Scale bar = 50 µm. Please click here to download this File.
Supplementary Figure 3: Characterization of MSCs and human endometrial stromal cells (ESCs) as the negative control. (A) MSCs were incubated with FITC-IgG and were negative for the FITC signal. (B) For adipogenic differentiation, there were no lipid droplets in ESCs. Scale bar = 50 µm. (C) For osteogenic differentiation, there were no calcium deposits in ESCs. Scale bar = 50 µm. (D) There was no NSE signal in ESCs. Scale bar = 50 µm. Please click here to download this File.
Supplementary Figure 4: Profiles of cell surface markers in the stem cells, and IgG. UCA-PSCs, UCV-PSCs, and WJ-MSCs were negative for CD34, CD45, and HLA-DR. UCA-PSCs, UCV-PSCs, and WJ-MSCs were positive for CD13 and CD73. IgG as overlay isotype control was negative for CD34, CD45, HLA-DR, CD13, and CD73. The results of three repeated experiments were analyzed, which proved the method was reproducible. The error bars represent the means ± SE of three independent experiments performed in triplicate. Please click here to download this File.
Discussion
This study isolated three different kinds of cells from the umbilical cord arteries, vein, and Wharton’s jelly. The umbilical cord was delivery waste, and its use was simple, safe, and without ethical dispute5. UC-MSCs are original and have strong differentiation ability1. Previous studies have shown that the amount of UC-MSCs isolated from umbilical cords by the collagenase, trypsin, and hyaluronidase digestion method was not abundant; the stem cells cannot be passaged many times in approximately 70% of the specimens which cannot meet clinical needs10. The other separation methods of UC-MSCs, such as flow cytometry or immunomagnetic bead techniques, were complex and expensive11. The adherent separation method6 can rapidly and massively isolate UC-MSCs. Their differentiation and proliferation ability are stronger than the cells isolated by collagenase digestion in vitro12. It can be expanded to obtain sufficient MSCs quickly to meet the needs of clinical treatment13,14.
The stem cells were isolated and cultured from the umbilical cord using the adherent method. The cells were isolated from the adherent tissue and adhered to the wall as spindle-like fibroblasts. The umbilical cord arteries, vein, and Wharton’s jelly were segmented to 1 cm and placed in a cell culture dish. The cell culture dish was inverted in a cell incubator for 3 h before the cell culture medium was added2. The critical steps in the protocol were the proper size of the umbilical cord arteries, vein, and Wharton’s jelly, and the time to invert the cell culture dish. The cell dish was inverted for 3 h to make the tissue block adhere closely. After adding the cell culture medium, the tissue block was still fixed on the cell dish, which is important for the subsequent cell crawling out5. Troubleshooting of the technique involved the failure of cells to crawl out, which was due to the umbilical cord tissue leaving the body for more than 24 h15.
The limitations of this technique consisted of the risk of cell culture contaminants and failure of cell crawling out due to a long period of time for the isolation and culture16. However, the method of the isolation and culture of the stem cells used was economical and could obtain a large number of cells, which had significance with respect to existing methods5. The detection of cell surface molecular markers showed that the cells did not express CD34, CD45, or HLA-DR, but highly expressed CD13 and CD73. MSCs can be induced to differentiate into various mesoderm cells under specific conditions17; this multidirectional differentiation ability is the basis for MSCs to exert their repair function18. In this study, UCA-PSCs, UCV-PSCs, and WJ-MSCs differentiated into osteoblasts, adipocytes, and neuron-like cells under the induction of osteogenesis, adipogenesis, and neurogenesis in vitro, which is in accordance with the definition of US-MSCs19. Studies have shown that MSCs derived from different regions of the umbilical cord possess different characterisation15,16, indicating the importance of isolating the stem cells.
In recent years, there has been a new understanding of the origin of MSCs. Almost all MSCs come from perivascular tissue19. One study found that pericytes exist around the vascular region and are mainly located in capillaries and microvessels13. Pericytes express MSC markers, have multidirectional differentiation ability, and are the progenitor cells of MSCs20. Pericytes are located on the basal side of endothelial cells21. Pericytes around the vascular endothelium form a close connection with the vascular basement membrane22. Studies have shown that pericytes are the progenitor cells of MSCs, which play an important role in maintaining vascular endothelial function and vascular integrity in injury repair23. Pericytes can secrete more angiogenic factors, such as fibroblast growth factor-2 and vascular endothelial growth factor, to promote angiogenesis24. Pericytes derived from the pancreas have osteogenic properties and significantly promote angiogenesis25, which promotes the growth, development, and repair of tissue damage26. The speed and quality of angiogenesis are key factors for damage repair27. The damaged tissue must obtain enough nutrients and oxygen from the blood supply to ensure cell migration, proliferation, and differentiation28. Three different sources of stem cells have different characteristics and play different roles in the application. The previous study showed that UCA-PSCs expressed the angiogenesis related genes, CD146 and Jagged1, and had better angiogenesis ability than UCV-PSCs and WJ-MSCs5, which indicated the future applications of the technique in therapy for ischemia29.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors wish to acknowledge support from the Basic Research Project of Changzhou science and Technology Bureau under grant number CJ20200110 (to YJY), the National Nature Science Foundation of China (82001629, XQS), the Youth Program of Natural Science Foundation of Jiangsu Province (BK20200116, XQS), and Jiangsu Province Postdoctoral Research Funding (2021K277B, XQS).
Materials
| Adipogenic differentiation kit | Gibco | A1007001 | Multidirectional differentiation |
| Alizarin red staining solution | Sigma | A5533 | Multidirectional differentiation |
| Antibody against CD13 | Thermo Fisher Scientific | MA1-12034 | flow analysis |
| Antibody against CD34 | BD Biosciences | 560942 | flow analysis |
| Antibody against CD45 | BD Biosciences | 561865 | flow analysis |
| Antibody against CD73 | BD Biosciences | 940294 | flow analysis |
| Antibody against HLA-DR | BD Biosciences | 555560 | flow analysis |
| Anti-fluorescence quenching agent | Abcam | AB103748 | Immunofluorescence |
| Anti-Mouse IgG H&L (Alexa Fluor 488) | abcam | ab150113 | Multidirectional differentiation |
| ATRA | STEMCELL Technologies | 302-79-4 | cell culture |
| bFGF | Gibco | 13256029 | Multidirectional differentiation |
| BSA | Sigma | V900933 | Immunofluorescence |
| Cell incubator | Thermo Fisher Scientific | HERAcell 240i | cell culture |
| Cell-counting kit-8 | Dojindo | CK04 | cell proliferation |
| Centrifuge | Thermo Fisher Scientific | Sorvall™ MTX-150 | cell culture |
| DAPI | Sigma | 10236276001 | Immunofluorescence |
| DMSO | Sigma | D1435 | cell culture |
| FBS | Gibco | 10099141 | cell culture |
| FITC Mouse Anti-Human IgG | BD Biosciences | 560952 | flow analysis |
| Flow Cytometer | Thermo Fisher Scientific | A24864 | flow analysis |
| Fluorescence microscope | Thermo Fisher Scientific | IM-5 | flow analysis |
| Gelatin | Sigma | 48722 | Multidirectional differentiation |
| Leica Microscope | Leica | DM500 | Multidirectional differentiation |
| LG-DMEM medium | Gibco | 11-885-084 | cell culture |
| Microplate reader | Thermo Fisher Scientific | A51119500C | cell proliferation |
| Neurogenic induction | Gibco | A1647801 | Multidirectional differentiation |
| Oil red O solution | Sigma | O1516 | Multidirectional differentiation |
| Osteogenic induction | Cyagen | HUXXC-90021 | Multidirectional differentiation |
| Paraformaldehyde | Sangon Biotech | 30525-89-4 | Immunofluorescence |
| Pasteur pipette | Biosharp | BS-XG-03L | cell culture |
| PBS (phosphate buffered saline) | Hyclone | SH30256.LS | cell culture |
| Penicillin streptomycin | Hyclone | SV30010 | cell culture |
| Primary antibody against NSE | Santa Cruz Biotechnology | sc-292097 | Multidirectional differentiation |
| SPSS 22.0 | IBM | SPSS 22.0 | Statistical analysis |
| The cell climbing sheets | CITOTEST Scientific | 80346-0910 | Multidirectional differentiation |
| TritonX-100 | Sangon Biotech | 9002-93-1 | Immunofluorescence |
| Trypsin | Gibco | 25300120 | cell culture |
References
- Zhang, Y., et al. Comparison of the biological characteristics of umbilical cord mesenchymal stem cells derived from the human heterosexual twins. Differentiation. 114, 1-12 (2020).
- Yang, Y., et al. Transplantation of umbilical cord-derived mesenchymal stem cells on a collagen scaffold improves ovarian function in a premature ovarian failure model of mice. In Vitro Cellular & Developmental Biology-Animal. 55 (4), 302-311 (2019).
- Sepulveda, R. V., et al. Canine umbilical cord perivascular tissue: A source of stem cells for therapy and research. Research in Veterinary Science. 129, 193-202 (2020).
- Ahmed, T. A., Shousha, W. G., Abdo, S. M., Mohamed, I. K., El-Badri, N. Human adipose-derived pericytes: biological characterization and reprogramming into induced pluripotent stem cells. Cellular Physiology and Biochemistry. 54 (2), 271-286 (2020).
- Xu, L., et al. Different angiogenic potentials of mesenchymal stem cells derived from umbilical artery, umbilical vein, and Wharton’s jelly. Stem Cells International. 2017, 3175748 (2017).
- Garikipati, V. N. S., et al. Isolation and characterization of mesenchymal stem cells from human fetus heart. PLoS ONE. 13 (2), -192244 (2018).
- Simon, L. V., Hashmi, M. F., Bragg, B. N. APGAR Score. StatPearls. , (2022).
- Adler, E. M., Gough, N. R., Blundon, J. A. Differentiation of PC12 cells. Science’s STKE. 2006 (351), (2006).
- Liang, X., et al. Bisphenol A and several derivatives exert neural toxicity in human neuron-like cells by decreasing neurite length. Food and Chemical Toxicology. 135, 111015 (2020).
- Salehinejad, P., et al. Comparison of different methods for the isolation of mesenchymal stem cells from human umbilical cord Wharton’s jelly. In Vitro Cellular & Developmental Biology Animal. 48 (2), 75-83 (2012).
- Ding, D. C., Chang, Y. H., Shyu, W. C., Lin, S. Z. Human umbilical cord mesenchymal stem cells: a new era for stem cell therapy. Cell Transplant. 24 (3), 339-347 (2015).
- Han, Y. F., et al. Optimization of human umbilical cord mesenchymal stem cell isolation and culture methods. Cytotechnology. 65 (5), 819-827 (2013).
- Esteves, C. L., et al. Equine mesenchymal stromal cells retain a pericyte-like phenotype. Stem Cells and Development. 26 (13), 964-972 (2017).
- Nagamura-Inoue, T., He, H. Umbilical cord-derived mesenchymal stem cells: Their advantages and potential clinical utility. World Journal of Stem Cells. 6 (2), 195-202 (2014).
- Mennan, C., et al. Isolation and characterisation of mesenchymal stem cells from different regions of the human umbilical cord. BioMed Research International. 2013, 916136 (2013).
- Carvalho, M. M., Teixeira, F. G., Reis, R. L., Sousa, N., Salgado, A. J. Mesenchymal stem cells in the umbilical cord: phenotypic characterization, secretome and applications in central nervous system regenerative medicine. Current Stem Cell Research & Therapy. 6 (3), 221-228 (2011).
- Harichandan, A., Sivasubramaniyan, K., Buhring, H. J. Prospective isolation and characterization of human bone marrow-derived MSCs. Advances in Biochemical Engineering/Biotechnology. 129, 1-17 (2013).
- Zhang, H., et al. Acceleration of fracture healing by overexpression of basic fibroblast growth factor in the mesenchymal stromal cells. STEM CELLS Translational Medicine. 6 (10), 1880-1893 (2017).
- Caplan, A. I. New MSC: MSCs as pericytes are Sentinels and gatekeepers. Journal of Orthopaedic Research. 35 (6), 1151-1159 (2017).
- Caplan, A. I. MSCs: the sentinel and safe-guards of injury. Journal of Cellular Physiology. 231 (7), 1413-1416 (2016).
- Tigges, U., Komatsu, M., Stallcup, W. B. Adventitial pericyte progenitor/mesenchymal stem cells participate in the restenotic response to arterial injury. Journal of Vascular Research. 50 (2), 134-144 (2013).
- Herrmann, M., et al. Pericyte plasticity-comparative investigation of the angiogenic and multilineage potential of pericytes from different human tissues. European Cells and Materials. 31, 236-249 (2016).
- Ahmed, T. A., El-Badri, N. Pericytes: The role of multipotent stem cells in vascular maintenance and regenerative medicine. Advances in Experimental Medicine and Biology. 1079, 69-86 (2018).
- Greenberg, J. I., et al. A role for VEGF as a negative regulator of pericyte function and vessel maturation. Nature. 456 (7223), 809-813 (2008).
- Lin, S. L., et al. Targeting endothelium-pericyte cross talk by inhibiting VEGF receptor signaling attenuates kidney microvascular rarefaction and fibrosis. American Journal of Pathology. 178 (2), 911-923 (2011).
- Failla, C. M., Carbo, M., Morea, V. Positive and negative regulation of angiogenesis by soluble vascular endothelial growth factor receptor-1. International Journal of Molecular Sciences. 19 (5), 1306 (2018).
- Kim, J. M., et al. Perivascular progenitor cells derived from human embryonic stem cells exhibit functional characteristics of pericytes and improve the retinal vasculature in a rodent model of diabetic retinopathy. STEM CELLS Translational Medicine. 5 (9), 1268-1276 (2016).
- Mills, S. J., Cowin, A. J., Kaur, P. Pericytes, mesenchymal stem cells and the wound healing process. Cells. 2 (3), 621-634 (2013).
- Dar, A., et al. Multipotent vasculogenic pericytes from human pluripotent stem cells promote recovery of murine ischemic limb. Circulation. 125 (1), 87-99 (2012).

.
