Retroviral CRISPR/Cas9-Mediated Gene Targeting for the Study of Th17 Differentiation in Vitro
Summary
We present a protocol for retroviral transduction of guide RNA into primary T cells from Cas9 transgenic mice, providing an efficient alternative for gene editing in studying Th17 differentiation.
Abstract
T helper cells that produce IL-17A, known as Th17 cells, play a critical role in immune defense and are implicated in autoimmune disorders. CD4 T cells can be stimulated with antigens and well-defined cytokine cocktails in vitro to mimic Th17 cell differentiation in vivo. Research has been conducted extensively on the Th17 differentiation regulation mechanisms using the in vitro Th17 polarization assay.
Conventional Th17 polarization methods typically involve obtaining naïve CD4 T cells from genetically modified mice to study the effects of specific genes on Th17 differentiation and function. These methods can be time-consuming and costly and may be influenced by cell-extrinsic factors from the knockout animals. Thus, a protocol using retroviral transduction of guide RNA to introduce gene knockout in CRISPR/Cas9 knockin primary mouse T cells serves as a very useful alternative approach. This paper presents a protocol to differentiate naïve primary T cells into Th17 cells following retroviral-mediated gene targeting, as well as the subsequent flow cytometry analysis methods for assaying infection and differentiation efficiency.
Introduction
Th17 cells, a unique subset of CD4+ T helper cells, are vital for eradicating extracellular bacteria and fungi and play a significant role in various autoimmune diseases1,2,3. Emerging evidence suggests that Th17 cells exhibit heterogeneity, functioning in both pathogenic and non-pathogenic conditions, influenced by environmental and genetic factors. Elucidating the regulatory processes that control the differentiation of Th17 cells, plasticity, and heterogeneity is crucial for the advancement of more effective immunotherapeutic strategies.
Genetically modified animals have been widely used to unveil the key regulators of Th17 cell differentiation and functions. Using genetically modified animals involves complete manipulation in vivo, providing authenticity and systematic study of its role in physiological conditions or disease models. Nevertheless, high-throughput screening with this approach is largely impractical. In vitro polarization assays provide an alternative for studying Th17 cell differentiation. Interleukin 6 (IL-6) in combination with transforming growth factor β1 (TGFβ1) has been shown to promote the development of non-pathogenic Th17 cells, while IL-6, IL-1β, and IL-23 are implicated in driving the differentiation of pathogenic Th17 cells (pTh17)4,5.
The emergence of CRISPR/Cas9 technology has facilitated precise genome editing at specific bases. When combined with retroviral transduction, this approach provides a potent, efficient, and economical genetic method for screening and functionally studying potential regulators in Th17 cells6,7. In this study, we improved the procedure for retroviral transduction within Th17 polarization system. Using a retroviral system, we infected pre-established Cas9-expressing activated naïve T cells from mice. The cells were transduced with guide RNA (gRNA) constructs driven by the U6 promoter, along with genes encoding fluorescent reporter proteins under the control of the EF1a promoter to facilitate the knockout of the target gene. Then, the transduced T cells were cultured under specific cytokine conditions to induce differentiation into Th17 cells. Notably, knocking out RoRγt significantly reduced IL-17A production compared to the control group. The effectiveness of this system depends on optimized retrovirus production and transduction conditions for activated primary T cells, providing a rapid and practical approach for studying specific genes in Th17 differentiation and function.
Protocol
All procedures were approved by the Experimental Animal Welfare Ethics Committee, Renji Hospital, Shanghai Jiao Tong University School of Medicine and are in compliance with institutional guidelines.
1. Retroviral production
- Preparation of retroviral construct
- Use the CRISPick tool (https://portals.broadinstitute.org/gppx/crispick/public) to design the sgRNA for RORγt knockout8. Specify the reference genome as Mouse GRCm38, and choose the PAM sequence as NGG (for SpyoCas9, Hsu (2013) tracrRNA) based on CRISPRko guide system. Validate the target gene's name or other target gene formats and submit the valid input to obtain sgRNA candidates. Setting other parameters to default values, set sgRNA target to RORγt (Rorc, Gene ID: 19885) to disturb the differentiation of Th17 and select non-targeting sgRNA (abbr. sgNT, sequence: GCGAGGTATTCGGCTCCGCG) from the mouse GeCKO v2 libraries.
NOTE: This tool offers a range of clickable options for designing guide RNAs, allowing for convenient customization to meet specific requirements.- Enter the target gene name in the search box under the Quick Gene Lookup column. The system will display the gene ID and the full name of the target gene to ensure accurate selection.
NOTE: Additional options are available for validating different target gene formats.
- Enter the target gene name in the search box under the Quick Gene Lookup column. The system will display the gene ID and the full name of the target gene to ensure accurate selection.
- Choose one sgRNA targeted on Rorc (abbr. sgRorc) sequence according to the high combined rank and pick order to avoid off-target matches and increase on-target efficacy (sgRorc sequence: GTCATCTGGGATCCACTACG). Synthesize two oligonucleotides for sgRorc and sgNT: for the forward oligo, append the sequence "gttttagagctagaaatagcaagttaaaat" to the 5' end of each guide sequence; for the reverse oligo, append the sequence "tttcgtcctttccacaagatatataaagc" to the 3' end of each guide sequence.
NOTE: Typically, multiple options exist for sgRNA sequences. It is essential to select 2 or 3 sgRNA sequences targeting the same genotype to assess their knockout efficiency. In our preliminary experiments, two gRNAs target to Rorc were selected for comparison (sgRNA sequence1: GTCATCTGGGATCCACTACG; sgRNA sequence2: CTTGAGTATAGTCCAGAACG). Experimental results indicated that the gRNA presented in the current method (sgRNA sequence1) exhibits higher knockout efficiency between the two. - Amplify the sgRorc and sgNT coding sequences using DNA polymerase in PCR with the above primers and clone them into pMX-U6-MCS vector (between the regions of U6 promoter and the gRNA scaffold) in frame with mCherry fluorescent protein, respectively9. Set up the reaction system (50 µL) as Table 1. Those primers consist of sgRNA sequence (3'), which can insert the sgRNA into the vector, and a segment of vector sequence (5'), which can amplify the full-length vector fragment. Additionally, design the forward and reverse primers with an overlap of approximately 18 bp within the sgRNA region (but not fully overlapping), facilitating the assembly of the linearized cloning vector during the ligation process.
NOTE: A linearized vector fragment at the site of sgRNA sequence with primers by PCR of approximately 5,551 kd should be obtained. - Purify the construct using a gel extraction kit and circularize it using a cloning kit. Use the recombination products for transformation assays.
- Pick the single clones from plates and verify sgRorc or sgNT-coding sequences via first-generation sequencing, using U6 promoter primer (sequence: ATGGACTATCATATGCTTACCGTA).
- Select the desired single clone for amplification in the LB broth. Extract high-quality plasmid DNA from bacterial cells by using an endotoxin-free plasmid kit.
NOTE: Make sure that plasmid solution has a high quality. We recommend that the concentration of plasmid solution should be at least 1 µg/µL.
- Use the CRISPick tool (https://portals.broadinstitute.org/gppx/crispick/public) to design the sgRNA for RORγt knockout8. Specify the reference genome as Mouse GRCm38, and choose the PAM sequence as NGG (for SpyoCas9, Hsu (2013) tracrRNA) based on CRISPRko guide system. Validate the target gene's name or other target gene formats and submit the valid input to obtain sgRNA candidates. Setting other parameters to default values, set sgRNA target to RORγt (Rorc, Gene ID: 19885) to disturb the differentiation of Th17 and select non-targeting sgRNA (abbr. sgNT, sequence: GCGAGGTATTCGGCTCCGCG) from the mouse GeCKO v2 libraries.
- Preparation of packaging cells
- Prepare Platinum-E (Plat-E) cells using the following procedure: culture Plat-E cells with Dulbecco's Modified Eagle's Medium (DMEM medium) containing 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 0.1 mg/mL streptomycin in a 10 cm culture dish. It will be referred to as the cell culture medium in the following.
NOTE: To obtain the proper transfection efficiency, we recommend using cells in the logarithmic growth phase with less than 15 passage numbers. - Split a full plate of 10 cm Plat-E cells (~2.6 × 107 cells) at a proper ratio into a new 10 cm dish 1 day before transfection to achieve a cell confluency of approximately 80% the next day, which is the optimal condition for transfection. Perform cell passaging for a new 10 cm dish at a 1:3 ratio (seed ~8 × 106 cells in a new 10 cm dish the day before transfection).
NOTE: The split ratio is contingent upon the growth condition of the cells, with higher transfection efficiency achievable in cells exhibiting robust growth. We perform cell passaging for a 10 cm dish at a 1:3 ratio. For different cell culture dishes, such as a 6 cm dish, we recommend that 1.5 × 106 Plat-E cells should be seeded the day before transfection.
- Prepare Platinum-E (Plat-E) cells using the following procedure: culture Plat-E cells with Dulbecco's Modified Eagle's Medium (DMEM medium) containing 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 0.1 mg/mL streptomycin in a 10 cm culture dish. It will be referred to as the cell culture medium in the following.
- Transfection
- One hour before transfection, substitute the cell culture medium with 8 mL of reduced serum medium without phenol red with 5% FBS, but without penicillin-streptomycin, as it may interfere with the transfection reagent. Warm the reduced serum medium to 37 °C before use.
- Prepare the transfection mixture containing mix 1 and mix 2: mix 1 includes 18 µg of plasmid DNA (pMXs-U6-MCS-sgNT-mCherry/pMXs-U6-MCS-sgRorc-mCherry) and 6 µg of pCL-Ecotropic Retroviral Vector (pCL-Eco) in 1,000 µL of reduced serum medium. Mix 2 contains 48 µL of transfection reagent in 1,000 µL of reduced serum medium. Add mix 2 drop by drop into mix 1 and mix gently and thoroughly. Incubate the complex for 15-20 min at 20-25 °C.
- Carefully drip the mixture into the Plat-E cells, swirling the plate gently to ensure even distribution. Incubate the cells at 37 °C under 5% CO2 for 6-12 h.
- Replace the medium with 10 mL of fresh cell culture medium and continue incubating the cells at 37 °C, 5% CO2 for 48 h.
- Assess the transfection efficiency in Plat-E cells by detecting the retroviral vector tag, serving as a proxy, via fluorescence microscopy or flow cytometry.
NOTE: A strong mCherry fluorescence (the fluorescence positive exceeds 80%) signal was detected in a considerable proportion of Plat-E cells transfected with the pMX-U6-MCS vector, indicating successful transfection. - Harvest the virus-containing culture supernatant and centrifuge at 1,500 × g for 10 min to remove cell fragments. Aliquot the virus supernatant 1 mL per vial and store at -80 °C.
NOTE: Filtration is an alternative method for removing cell debris. We recommend using a syringe filter with a 0.45 µm pore size and a polyethersulfone (PES) membrane to minimize the retention of viral particles on the filter membrane.
2. Retroviral infection of activated CD4 + T cells and Th17 differentiation
- Prepare and activate primary naïve CD4+ T cells
- Coat each well of a 24-well cell culture plate with 2 µg/mL of anti-CD3e and 4 µg/mL of anti-CD28 in sterile 500 µL PBS, wrap the plate in parafilm to prevent evaporation and contamination, and then incubate plate at 4 °C overnight (or 37 °C for 1-2 hours before plating CD4+ T cells) at the day before isolating naïve CD4+ T cells.
NOTE: This step is needed to perform plate-bound stimulation for the mouse T cells before performing viral transduction. - On the next day, euthanize 6-8-week-old Cas9-expressing mice (R26-CAG-Cas9 mice) by CO2 asphyxiation and sterilize them with 70% ethanol.
- Harvest the mouse spleen and place it in PBS with 2% FBS and 100 µM EDTA (FACS buffer).
- Grind the spleen with sterilized ground glass (matte side) in the FACS buffer, homogenize the tissue into a single-cell suspension, and filter the cell suspension through a 70 µm cell strainer into a 50 mL tube.
NOTE: Be sure to grind the tissue gently and consistently to keep the tissue moist. Violently grinding the tissue can affect cell survival dramatically. - Spin the splenic cell suspension at 800 × g for 5 min. Remove the supernatant and resuspend the cell pellet in 2 mL of red cell lysis buffer. Let it sit at room temperature for 5 min, then add FACS buffer to reach a total volume of 15 mL. Centrifuge the splenocytes at 800 × g for 5 min.
- Resuspend the splenocytes and filter them through a 40 µm cell strainer. Adjust the final volume to 1 mL for the CD4+ T cell isolation.
- Isolate CD4+ T cells using commercial reagent kits by following the instructions in the manual.
- Label the CD4+ T cells with fluorescent antibodies mixture (CD4, CD25, CD44, and CD62L) at 4 °C for 30 min, wash the cells with FACS buffer, and isolate the naïve CD4+ T cells using a flow cell sorter. This will result in obtaining CD4+CD25–CD62LhighCD44low naïve T cells.
- Remove the plate-bound stimulation supernatant from the 24-well cell culture plate (prepared in step 2.1.1) and rinse each well twice with 500 µL of PBS. Plate 1 x 106 naïve CD4+ T cells in 1 mL of lymphocyte culture medium in a well of the 24-well cell culture plate. Incubate the cells in the cell incubator.
NOTE: Lymphocyte culture medium contains 10% FBS, 100 units/mL penicillin, and 0.1 mg/mL streptomycin, 1 mM sodium pyruvate, 10 mM HEPES, and 50 µM β-ME in RPMI 1640 medium.
- Coat each well of a 24-well cell culture plate with 2 µg/mL of anti-CD3e and 4 µg/mL of anti-CD28 in sterile 500 µL PBS, wrap the plate in parafilm to prevent evaporation and contamination, and then incubate plate at 4 °C overnight (or 37 °C for 1-2 hours before plating CD4+ T cells) at the day before isolating naïve CD4+ T cells.
- Retroviral infection
- After 18-24 h, collect each well of activated cells into the 1.5 mL tube. Centrifuge at 800 × g for 5 min, and discard the supernatant.
- Resuspend the cell pellet in 1 mL of infection mixture (10 µg/mL hexadimethrine bromide, 10 mM HEPES in virus supernatant) and add 1 mL of the mixture to a new well of a 48-well cell culture plate. In the meanwhile, save enough cells as negative (un-transduced) control.
NOTE: It is feasible to have fewer than 1 x 106 activated naïve CD4+ T cells per well, but it is not recommended to use less than 0.5 x 106 cells per well. Additionally, no cells should be seeded in the peripheral wells of the 48-well culture plate. - Wrap the plate with parafilm, and centrifuge at 800 x g for 90 min at 32 °C. Set accelerate and decelerate at 3. After centrifugation, remove the parafilm, incubate the cells in the incubator for 4 h, and proceed with the differentiation steps.
- Differentiation of infected cells into Th17 cells
- Prepare the antigen-presenting cells (APC) during retroviral infection. First, obtain splenocytes from wild-type mice following the instructions from steps 2.1.2 to 2.1.4. Resuspend the splenocytes with 1 mL of lymphocyte culture medium with 50 µg/mL mitomycin in a 15 mL tube. Incubate the splenocytes at 37 °C under 5% CO2 for 1 h.
- One hour later, add PBS up to the 15 mL mark to wash the splenocytes, centrifuge at 800 x g for 5 min, discard the supernatant, resuspend the splenocytes with 1 mL of lymphocyte culture medium, and count the cell number. APC cells have been successfully prepared.
- After the 4 h of cell incubation (as in step 2.2.3), collect transduced CD4+ T cells in 1.5 mL tubes and centrifuge at 800 x g for 5 min. Discard the supernatant and resuspend infected cells in 600 µL of PBS to wash. Centrifuge at 800 x g for 5 min and discard the supernatant.
- Place 0.5 × 106 transduced CD4+ T cells in 1 mL of lymphocyte culture medium with 1.5 x 106 APC cells in a well of a 24-well cell culture plate. Supplement with 2 µg/mL of anti-CD3e, 2 µg/mL of anti-CD28, and Th17 cell differentiation cocktail, which contains 10 µg/mL anti-IFNγ, 10 µg/mL anti-IL-12/IL23 p40, 10 µg/mL anti-IL4, 2.5-3 ng/mL hTGF-β, and 20-30 ng/mL IL-6. Culture cells for 2 days.
- Collect each well of cells into the 1.5 mL tubes, centrifuge at 800 x g for 5 min, and discard the supernatant. Refresh new 1 mL of the lymphocyte culture medium only with 3 ng/mL TGF-β and 30 ng/mL IL-6. Place the cells in a new well of 24-well cell culture plate for an extended culture period. Analyze the cells 2 days after the medium change.
3. Evaluation of transduction efficiency and differentiation results
- After two days of medium change, collect cells and centrifuge at 800 × g for 5 min and discard the supernatant. Treat differentiated cells with 50 ng/mL phorbol 12-myristate 13-acetate (PMA), 500 ng/mL ionomycin and 5 µg/mL brefeldin A in 500 µL of lymphocyte culture medium in a new well of 24-well cell culture plate at 37 °C incubator for 4 h. Harvest half of the cells for flow cytometry analysis and lyse the other half of the cells with RNA extraction buffer for qPCR analysis.
- For flow cytometry analysis
- Collect the cells into 1.5 mL tubes. Add 1 mL of FACS buffer into tubes and centrifuge at 800 × g for 5 min to wash the cells.
- Resuspend the cell pellet with fluorescent antibodies mixture (CD4 and Fixable Viability Dye in 100 µL of PBS) and stain the cells at 4 °C for 30 min.
- Wash the cells with 300 µL of PBS and fix them with 300 µL of 2% phosphate-buffered formaldehyde (FA) at 4 °C for at least 60 min.
- One hour later, wash the cells with PBS, resuspend them with 600 µL of 1x permeabilization buffer, and centrifuge at 800 × g for 5 min.
- Discard the supernatant and stain with anti-IL-17A in 100 µL of 1x permeabilization buffer for 60 min at room temperature or 4 °C overnight.
- The next day, wash the cells with 1x permeabilization buffer and resuspend them in 200 µL of PBS. Analyze the cells by flow cytometry10. See the Representative Results section.
- For qPCR analysis, extract RNA following the instructions of the RNA extraction kit. Reverse-transcribe the RNA into cDNA for storage or subsequent qPCR analysis. The design of these primers should adhere to general qPCR primer design principles while also specifically targeting the region spanning the expected indel site, where gRNA-mediated cleavage has occurred. Any reduction in amplification efficiency or loss of signal at this site may serve as an indicator of successful knockout. The qPCR primers are as follows:
Rorc F: TCCACTACGGGGTTATCACCT; R: AGTAGGCCACATTACACTGCT.
IL17a F: TCAGCGTGTCCAAACACTGAG; R: CGCCAAGGGAGTTAAAGACTT.
Representative Results
In the study, we cloned the sgRNA target to Rorc and sgRNA-non-targeting coding sequences into pMX-U6-MCS vector with mCherry fluorescent protein (Figure 1A,B). Retrovirus production was carried out according to the protocol outlined in Figure 2. The transfection was initiated on day 0, and the retroviral harvest occurred on day 2. Transfection efficiency can be tested by the mCherry fluorescence intensity (Figure 1C). Before harvesting the virus, naïve CD4+ T cells were sorted and activated. These activated CD4+ T cells were then infected using spin infection, followed by continuation of the differentiation steps as described. Finally, the efficiency of transduction and the results of differentiation were evaluated using flow cytometry analysis and qPCR analysis.
Figure 3 illustrates the gating strategy applied for sorting naïve CD4+ T cells. After selecting the main cell population and singlet cells (as shown in the first two gates of Figure 3), FVD– CD4+ cells were gated to distinguish CD4 T cells in polarized to Th17 cells from those in APCs. Finally, IL-17A produced by CD4 T cells represent Th17 differentiation efficiency, which indicated the knockout efficiency. The percentage of mCherry-positive cells reflects the efficiency of retroviral infection, while the knockout efficiency is indicated by the percentage of IL-17A-positive cells. To enhance cell viability and minimize sorting duration, splenocytes were enriched using a mouse CD4+ T cell isolation kit, resulting in an enrichment efficiency surpassing 90%.
The efficiency of retrovirus infection, indicated by the presence of mCherry-positive cells, is typically around 40% (Figure 4A). Although infection efficiency can be increased through virus concentration, this approach comes at the expense of prolonged experimental duration and higher costs. In our experiments, infected cells cultured without the Th17 cell differentiation cocktail served as the Th17 differentiation negative control, designated as Th0. A non-targeting sgRNA was used as a positive control, referred to as sgNT. For gene editing, we employed gRNA targeting the RORγt gene (sgRNA-Rorc), which significantly reduced Th17 differentiation efficiency (Figure 4A). This intervention also led to a substantial decrease in the expression levels of Rorc and IL-17a (Figure 4B).
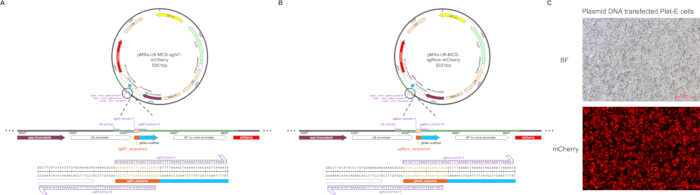
Figure 1: Example of sgRNA sub-cloned into a retroviral vector and transfection efficiency. The pMX-U6-MCS vector was used as the initial construct. The gRNA scaffold, along with the sgRNA sequences (the orange part) targeting the (A) scrambled site and (B) Rorc, were inserted downstream of the U6 promoter (the green part as a section for sequencing with U6 primers). The locations of the sgRNA-containing primers and sequencing primer sites are indicated below the circular vector The full names of the abbreviations on the plasmid are as follows. Abbreviations: AmpR = resistance to ampicillin, carbenicillin, and related antibiotics; LTR = long terminal repeat; MMLVψ = packaging signal of Moloney murine leukemia virus; gag = truncated Moloney murine leukemia virus (MMLV) gag gene lacking the start codon. (C) Microscopy imaging of mCherry in plasmid DNA-transfected Plat-E cells. Scale bar: 500 px. Please click here to view a larger version of this figure.
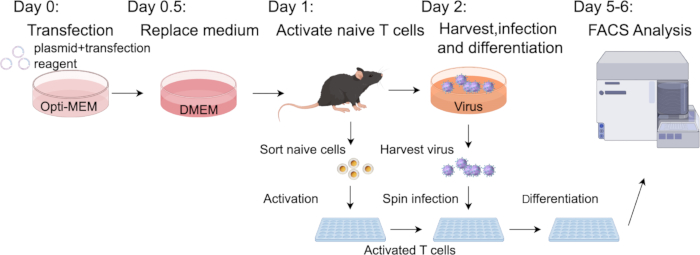
Figure 2: Schematic illustration of the protocol. The day before transfection, the density of the Plat-E cells needs to be adjusted to a proper range. The next day (day 0), perform transfection and replace medium 12 h later. On day 1, sort and activate the naïve T cells from Cas9-expressing mice. Subsequently (day 2), harvest the virus-containing culture supernatant and combine with the activated T cells for retroviral transduction. The infected cells are then differentiated into Th17 cells for 3–4 days and analyzed using FACS. Please click here to view a larger version of this figure.
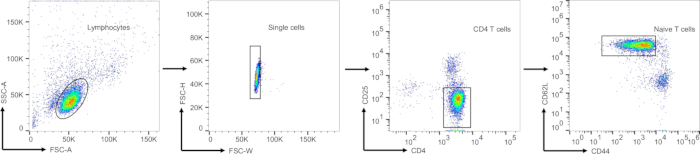
Figure 3: Gating strategy for sorting the CD4 naïve T cells. Surface staining was performed on the enriched CD4 T cells to label CD4, CD25, CD44, and CD62L. The gating strategy employed was as follows: initially, lymphocytes were gated based on forward scatter and side scatter parameters. Subsequently, single cells were identified using FSC-H and FSC-W. CD4+CD25– cells were then gated to exclude regulatory T cells (Tregs). Finally, CD44lowCD62Lhigh naïve T cells were identified. The isolated CD4+ naïve T cells were collected for subsequent experiments. Abbreviations: FSC = forward scatter; SSC = side scatter. Please click here to view a larger version of this figure.
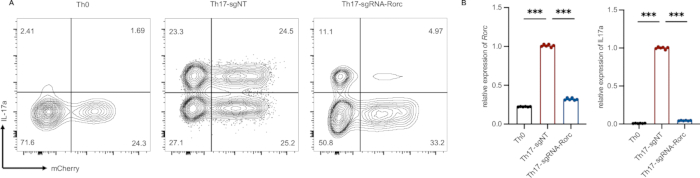
Figure 4: Example of successful transduction of activated T cells and Th17 differentiation. (A) The differentiation efficiency of Th17 is reduced significantly which is edited by RORγt gRNA (gating on mCherry+ IL-17a+population). (B) Rorγt and IL-17a relative mRNA expression in Th0 (infected cells cultured without Th17 cell differentiation cocktail), sgNT (non-targeting sgRNA) and sgRNA-Rorc (sgRNA targeting the RORγt gene). ***p < 0.001 (unpaired two-tailed student’s t-test; data are presented as mean ± SEM).Please click here to view a larger version of this figure.
| PCR Reaction Components | PCR Program | ||||
| Component | Volume (μL) | Step | Temp. | Time | Cycle |
| 2 × Phanta Mix (Dye Plus) | 25 | 1 | 95 ºC | 3 min | 1 |
| ddH20 | 20 | 2 | 95 ºC | 15 s | |
| Primer-F (10 μM) | 2 | 3 | 56 ºC | 15 s | |
| Primer-R (10 μM) | 2 | 4 | 72 ºC | 5 min 30 s | Go to Step 2 |
| 35 × | |||||
| Template (~30 ng) | 1 | 5 | 72 ºC | 5 min | 1 |
| 6 | 12 ºC | ∞ | |||
Table 1: PCR Reaction Components and PCR Program for gRNA vector construction.
Discussion
CRISPR/Cas9 genome editing via retroviral delivery is a robust method for exploring the roles of helper T cells. This protocol offers a rapid and effective approach to examining specific genes involved in Th17 differentiation and function. Several critical steps must be carefully followed to achieve optimal results. First, for enhanced gene knockout efficiency, gRNAs should be carefully selected. Given the risk of off-target effects in CRISPR/Cas9 gene editing, it is prudent to choose 2-3 gRNAs with high scores, as identified in protocol section 1.1.1, for further experiments11,12. Second, a high viral titer is essential for successful infection. Proper maintenance of the Plat-E packaging cell line is crucial. Cells should reach approximately 80% confluence on the day of transfection, as both inadequate and excessive cell numbers will compromise virus titers. If Plat-E cells are unavailable, co-transfecting Human Embryonic Kidney 293T cells with the pCL-Eco vector can also produce high viral titers13,14. Maintaining a high plasmid concentration of at least 1 µg/µL is important for achieving a high virus titer. It is essential to harvest the virus suspension before it turns lemon yellow since the virus exhibits sensitivity to pH alteration. Therefore, the culture medium's pH should be carefully regulated within an appropriate range. The virus should be stored at -80 ºC to maintain its integrity and protected from repeated freeze-thaw cycles15.
Third, to achieve high infection efficiency, primary cells must be adequately activated. Inadequate cellular activation significantly compromises viral infection efficiency16. Before viral infection, the activation status of T cells should be confirmed microscopically, as activated cells exhibit a notably larger size than inactivated cells. It is worth noting that our Th17 differentiation protocol has been specifically optimized for C57BL/6 mice, so adjustments may be necessary for other mouse strains. Fourth, to achieve the desired Th17 differentiation results, maintaining optimal cellular conditions is crucial. During spin infection, it is important to place the cells in the wells near the center to help maintain cell viability. Following spin infection, incubating the cells for 4 h (protocol step 2.2.4) is crucial for their recovery. Additionally, when transferring cells, gentle pipetting is important to maintain cell viability. Different stimulation conditions can impact Th17 differentiation. In this experiment, incorporating antigen-presenting cells into the differentiation system significantly enhanced differentiation efficiency compared to anti-CD3e and anti-CD28 plate-bound stimulation. Finally, it is crucial to ensure that all cytokines used remain functional during transport and storage. Cytokines can rapidly lose stability in repeated freeze-thaw cycles or inappropriate reconstitution solutions. Cold treatment and equal aliquots are necessary for cytokines to retain functions throughout the procedures.
This protocol for generating specific gene knockout Th17 cells does have several limitations. First, it is difficult to achieve complete transduction efficiency in primary mouse T cells. Some cells may not undergo the intended gene knockout during Th17 differentiation. Second, primary T cells have a limited lifespan in vitro. After approximately a week, these cells tend to die, posing challenges for subsequent experiments. Third, our protocol relies on Cas9 transgenic mice and when we attempted to incorporate the Cas9 sequence into the core plasmid, this resulted in a significant reduction in viral titers. Lastly, we notice that this protocol is not suitable for analyzing functions of genes involved in the early stages of naïve CD4+ T cell activation, as those cells must be activated prior to retroviral transduction.
Despite its limitations, this protocol has been successfully applied in various studies, facilitating the exploration of key genetic regulators involved in Th17 differentiation. It enables the assessment of changes in Th17 differentiation efficiency following the knockout of target genes in activated T cells. Additionally, this protocol is suitable for evaluating other aspects of Th17 cell function, including cell proliferation, survival, transcription factor stability, cytokine secretion, and immune responses. Moreover, our lab has demonstrated its effectiveness in the retroviral transduction and differentiation of induced regulatory T cells (iTregs). Although not all types of T cells have been tested, this protocol may also be applicable in other subsets, such as Th1, Th2, and CD8+ T cells. Overall, this technique serves as a valuable tool for conducting gene knockouts in immune cells, offering significant potential for uncovering critical genetic signals that regulate immune functions.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
We acknowledge Dou Liu, Dongliang Xu, and Pinpin Hou from the core facility of the Shanghai Immune Therapy Institute for their support in utilizing the instruments. This work was supported by National Natural Science Foundation of China Grants 31930038, U21A20199, 32100718, and 32350007(to Linrong Lu); 32100718 (to Xuexiao Jin); Innovative research team of high-level local universities in Shanghai SHSMU-ZLCX 20211600 (to Linrong Lu); Internal Incubation Program RJTJ24-QN-076(to Zejin Cui). Figure 2 was prepared with Figdraw.
Materials
| 0.5 M EDTA (pH 8.0) | Solarbio | E1170 | |
| 100 mm cell and tissue Culture Dish | BIOFIL | TCD010100 | |
| 1 M Hepes (Free Acid, sterile) | Solarbio | H1090 | |
| 24-well cell and tissue culture plate | NEST | 702002 | |
| 48-well cell and tissue culture plate | NEST | 748002 | |
| Brefeldin A Solution (1,000x) | BioLegend | 420601 | |
| Brilliant Violet 650 anti-mouse CD4 Antibody (RM4-5) | BioLegend | 100546 | |
| CD3e Monoclonal Antibody (145-2C11), Functional Grade, eBioscience | Invitrogen | 16-0031-82 | |
| CD44 Monoclonal Antibody (IM7), PE, eBioscience | Invitrogen | 12-0441-83 | |
| CD62L (L-Selectin) Monoclonal Antibody (MEL-14), APC, eBioscience | Invitrogen | 17-0621-82 | |
| Cell counter | Nexcelom Bioscience | Cellometer Auto 2000 | |
| Cell Strainer (40 μm) | biosharp | BS-40-CS | |
| Cell Strainer (70 μm) | biosharp | BS-70-CS | |
| Centrifuge | eppendorf | 5425 R | |
| Centrifuge | eppendorf | 5810 R | |
| ChamQ SYBR Color qPCR Master Mix | Vazyme | Q411-02 | |
| ClonExpress II One Step Cloning Kit | Vazyme | C112 | |
| DH5α Competent Cells | Sangon Biotech | B528413 | |
| Direct-zol RNA Miniprep | ZYMO RESEARCH | R2050 | |
| DMEM Medium | BasalMedia | L110KJ | |
| EasySep Mouse CD4+ T Cell Isolation Kit | STEMCELL | 19852 | |
| eBioscience Fixable Viability Dye eFluor 660 | Invitrogen | 65-0864-18 | |
| EndoFree Mini Plasmid Kit II | TIANGEN | DP118-02 | |
| ExFect Transfection Reagent | Vazyme | T101-01 | |
| Fetal Bovine Serum, Premium Plus | Gibco | A5669701 | |
| FITC anti-mouse IL-17A Antibody (TC11-18H10.1) | BioLegend | 506907 | |
| Formaldehyde solution | Macklin | F864792 | |
| HiScript IV RT SuperMix for qPCR(+gDNA wiper) | Vazyme | R423-01 | |
| Ionomycin | Beyotime | S1672 | |
| Mitomycin C | Maokang Biotechnology | 7/7/1950 | |
| Mouse GRCm38 | NCBI | RefSeq v.108.20200622 | |
| OPTI-MEM Reduced Serum Medium | Gibco | 31985070 | reduced serum medium |
| Pacific Blue anti-mouse CD4 Antibody (RM4-5) | BioLegend | 100531 | |
| PE/Cyanine7 anti-mouse CD25 Antibody (PC61) | BioLegend | 102015 | |
| PE/Cyanine7 anti-mouse CD4 Antibody (GK1.5) | BioLegend | 100422 | |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | 15140122 | |
| PMA/TPA | Beyotime | S1819 | |
| R26-CAG-Cas9 mice | Shanghai Model Organisms Center | Cat. NO. NM-KI-00120 | |
| Recombinant Human TGF-beta 1 (CHO-Expressed) Protein, CF | R&D Systems | 11409-BH | |
| Recombinant Murine IL-6 | PeproTech | 216-16 | |
| Research Cell Analyzer | BD Biosciences | BD LSRFortessa | |
| Research Cell Sorter | SONY | MA900 | |
| RPMI 1640 Medium | BasalMedia | L210KJ | |
| SimpliAmp Thermal Cycler PCR System | Applied Biosystems | A24811 | |
| Sodium pyruvate solution (100 mM) | Sigma-Aldrich | S8636 | |
| Ultra-LEAF Purified anti-mouse CD28 Antibody (37.51) | BioLegend | 102121 | |
| Ultra-LEAF Purified anti-mouse IFN-γ Antibody (XMG1.2) | BioLegend | 505847 | |
| Ultra-LEAF Purified anti-mouse IL-4 Antibody (11B11) | BioLegend | 504135 | |
| Ultra-LEAF Purified anti-mouse IL-12/IL-23 p40 Antibody (C17.8) | BioLegend | 505309 | |
| β-Mercaptoethanol (50 mM) | Solarbio | M8211 |
Referencias
- Wu, B., Wan, Y. Molecular control of pathogenic th17 cells in autoimmune diseases. Int Immunopharmacol. 80, 106187 (2020).
- Dong, C. Th 17 cells in development:: An updated view of their molecular identity and genetic programming. Nat Rev Immunol. 8 (5), 337-348 (2008).
- Patel, D. D., Kuchroo, V. K. Th17 cell pathway in human immunity: Lessons from genetics and therapeutic interventions. Immunity. 43 (6), 1040-1051 (2015).
- Ghoreschi, K., et al. Generation of pathogenic Th17 cells in the absence of TGF-β signalling. Nature. 467 (7318), 967-U144 (2010).
- Lee, Y., et al. Induction and molecular signature of pathogenic th17 cells. Nat Immunol. 13 (10), 991-999 (2012).
- Yang, H., et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 154 (6), 1370-1379 (2013).
- Yang, H., Wang, H., Jaenisch, R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc. 9 (8), 1956-1968 (2014).
- Doench, J. G., et al. Optimized sgrna design to maximize activity and minimize off-target effects of crispr-cas9. Nat Biotechnol. 34 (2), 184-191 (2016).
- Holst, J., et al. Generation of t-cell receptor retrogenic mice. Nat Protoc. 1 (1), 406-417 (2006).
- Ordonez, P., Bishop, K. N., Stoye, J. P., Groom, H. C. T. Analysis of SAMHD1 restriction by flow cytometry in human myeloid U937 cells. J Vis Exp. (172), (2021).
- Akcakaya, P., et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 561 (7723), 416-419 (2018).
- Bae, S., Park, J., Kim, J. -. S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of cas9 rna-guided endonucleases. Bioinformatics. 30 (10), 1473-1475 (2014).
- Singh, Y., Garden, O. A., Lang, F., Cobb, B. S. Retroviral transduction of helper T cells as a genetic approach to study mechanisms controlling their differentiation and function. J Vis Exp. (117), e54698 (2016).
- Naviaux, R. K., Costanzi, E., Haas, M., Verma, I. M. The pCL vector system: Rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 70 (8), 5701-5705 (1996).
- Cante-Barrett, K., et al. Lentiviral gene transfer into human and murine hematopoietic stem cells: Size matters. BMC Res Notes. 9, 312-312 (2016).
- Zhong, S., Malecek, K., Perez-Garcia, A., Krogsgaard, M. Retroviral transduction of T-cell receptors in mouse T-cells. J Vis Exp. (44), 2307 (2010).

.
