Label-Free Non-Linear Optics for the Study of Tubulin-Dependent Defects in Central Myelin
Summary
In this article, we present a protocol to detect microtubule-loaded oligodendrocytes in a model of tubulinopathy through a simple, innovative second harmonic generation microscopy approach.
Abstract
The satisfactory visualization of cytoskeletal components in the brain is challenging. The ubiquitous distribution of the networks of microtubules, microfilaments, and intermediate filaments in all the neural tissues, together with the variability in the outcomes of fluorescent protein fusion strategies and their limited applicability to dynamic studies of antibodies and drugs as chromophore vehicles, make classical optical approaches not as effective as for other proteins. When tubulin needs to be studied, the label-free generation of second harmonics is a very suitable option due to the non-centrosymmetric organization of the molecule. This technique, when conjugated to microscopy, can qualitatively describe the volumetric distribution of parallel bundles of microtubules in biological samples, with the additional advantage of working with fresh tissues that are unfixed and unpermeabilized. This work describes how to image tubulin with a commercial second harmonic generation microscopy setup to highlight microtubules in the tubulin-enriched structures of the oligodendrocytes, as in hypomyelination with atrophy of the basal ganglia and cerebellum (H-ABC) tubulinopathy, a recently described myelin disorder.
Introduction
The optical imaging of cytoskeletal structures in tissues and organ preparations is not an easy task. Cytoskeletal filaments are ubiquitous, so if generic staining is performed, for example, against alpha-tubulin or beta-actin or potentially keratin in an epithelial sample, the signal will likely be distributed rather homogeneously all over the sample. To restrict the staining to a more meaningful subset of cellular components, one can either use transgenic mice with targeted expression1 or plan to use isoform-specific antibodies. While very few of the latter are on the market (and very few exist at all2,3,4), a transgenic animal model might be available. However, it needs to be acquired by the lab and properly housed, with all the expenses involved in the process. Certain antibodies or chemicals, for example, fluorophore-conjugated drugs like phalloidin or paclitaxel, may be partially or fully incompatible with use in living cells or tissues, thus limiting their applicability to only studies of fixed samples.
In the case of tubulin, an additional aspect has to be taken into consideration, which is the sensitivity of the polymer to fixation. Conventional chemical fixation with formaldehyde is known for not being adequate for optimally preserving the integrity of microtubules5. Additionally, a recent report confirms that formaldehyde crosslinking induces subtle changes in the ultrastructure of the microtubule, similar to what happens with the binding of some drugs or physiological molecules such as GTP6.
The direct visualization of microtubules in unstained, unfixed samples is, therefore, often desirable. To achieve this, one technical solution is second harmonic generation (SHG) microscopy7, which is based on the ability of bundles of parallel microtubules to act as harmonophores and to emit frequency-doubled light when properly illuminated with an intense, pulsed infrared laser. Although a stronger and more stable second harmonic signal can be generated from collagen and myosin, which are the only other two biological materials known to be capable of frequency-doubling, the signal from tubulin has been used so far mostly to study mitotic spindle rearrangements8,9,10 and axonal microtubule morphology11,12,13.
In this work, we introduce a novel use of SHG microscopy as a diagnostic tool to distinguish central nervous system (CNS) tissues affected by tubulin beta 4 A (TUBB4A) tubulinopathy from their healthy counterparts14. Some of the mutations occurring in this predominantly neural isoform of tubulin, like those causing hypomyelination and atrophy of the basal ganglia and cerebellum (H-ABC), induce microtubule overfilling in the oligodendrocytes15,16; the cytoskeletal alterations, in turn, are associated with downstream effects like dysmyelination, with profound impairment of the motor and sensory pathways16,17,18,19. The taiep murine model used in this work displays abnormal microtubule content in the oligodendrocytes and recapitulates most of the sensory-motor symptoms of H-ABC patients17. The protocol explains how to image structures as the corpus callosum and the cerebellum, which are usually highly myelinated and which are severely affected in human patients as well as in the taiep rat19, to highlight the differences in SH signals between healthy and mutant tissues.
Protocol
All procedures described were done in compliance with the laws and codes approved in the seventh title of the Regulation of the General Health Law Regarding Health Research of the Mexican Government (NOM-062-ZOO-1999) and in accordance with the recommendations of the National Institutes of Health Guide for the Care and Use of Experimental Animals and were approved by the institutional committee of bioethics in research of the Universidad de Guanajuato and Benemérita Universidad Autónoma de Puebla.
1. Microscope settings
- Switch on the microscopy system.
- Turn on the pulsed laser to guarantee that it will be ready to lase at an optimal and steady power level after the sample extraction and preparation.
- Tune the laser to 810 nm. To study tissular microtubules, 10%-20% of the available laser power is used, which, in the described system, corresponds to 13-26 mW measured at the back focal plane of the objective.
- Make sure the microscope is Köhler-aligned (Supplementary File 1), with the objective that will be used for the SHG imaging.
NOTE: This is important since, in the commercial setup used in this work, the detection is performed in transmission mode and through the condenser. - Remove unwanted filters from the optical detection path (Figure 1A).
- Make sure the condenser diaphragm is fully opened to ensure that no light is unnecessarily stopped at this level.
- Prepare a 25x/0.8 numerical aperture (NA) oil-immersion objective with a small drop of immersion oil in the lens.
NOTE: This objective was used to best match the condenser NA, which was 0.55 (ideally, NAcond≥ NAobj).
2. Microscope preliminary controls
NOTE: Perform the preliminary microscope controls once, unless the setup is modified.
- Image dry corn starch sandwiched between glass slides with SHG parameters to generate SHG images that reveal the laser polarization direction. Follow the steps reported in Supplementary File 2, except for the laser power, which is less than 5% for corn starch.
- Take one image of sparse corn starch grains, and mark the orientation of the SHG lobules in the x-y plane. This orientation corresponds to the laser orientation (Figure 2A).
- As a control, take another image of the same sample, inserting into the optical path a half-wave plate (position shown in Figure 1A) to alter the oscillation direction of the laser. The resulting image displays rotated SHG signal lobules (Figure 2B, C).
- If using a different type of bandpass filter for the SHG, make sure it has optimal transmission properties by comparing the images (Figure 2D, E) and the signal intensities pixel by pixel along a line-scan (Figure 2F).
3. Tissue extraction
NOTE: Always use clean tools to perform the surgical procedures.
- Use 6-12 month old taiep rats and age-matched Sprague-Dawley controls (WT).
- Anesthetize the animals with an i.p. injection of a mixture of ketamine-xylazine (0.125 mg/Kg and 5 mg/Kg) diluted in a 0.9% sterile NaCl solution.
- Check the pain reflexes, and proceed only if they are absent.
- Sacrifice the animal via decapitation.
- Once the head is isolated, open the bones of the cranial vault carefully along the midline, starting from the most caudal point at the top toward the nose, from the orbital cavities toward the midline, and then from the most caudal point to below the brain and cerebellum.
NOTE: Bone cutters and Rongeurs allow for proper removal of the head bones without damaging the brain structures. - Release the brain and cerebellum from the bone. The two hemispheres could be separated. If the hemispheres are divided, use one half for SHG imaging, and fix the other half in 3.7% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min inside a chemical hood for subsequent sectioning and staining.
4. Vibratome sectioning
- Prepare a vibratome buffer tray filled with warm (37 °C) Hank's Balanced Salt Solution (HBSS).
- Fix the brain to the specimen plate using cyanoacrylate glue via a piece of masking tape. The most caudal portion/region contacts the glue so that the usable coronal sections from the opposite, rostral portion can be cut.
- Allow ~10 s for the glue to polymerize to obtain effective attachment of the sample.
NOTE: The best results are obtained when the brain is oriented so that the blade cuts "from top to bottom" rather than "from side to side" (Figure 1B).
- Allow ~10 s for the glue to polymerize to obtain effective attachment of the sample.
- Transfer the specimen plate onto its magnetic support inside the buffer tray.
- Start cutting 300-500 µm sections until the slices produced encompass the entire surface of the brain.
- At this point, reduce the thickness of the section to 160 µm.
- Once a complete 160 µm section is cut at the level of the corpus callosum, recover it with a modified glass Pasteur pipette with a large, flamed orifice (Figure 1C).
- Transfer it to a clean Petri dish with new warm HBSS or directly onto the glass support for microscopy (coverslip or glass-bottom dish).
NOTE: For the described experimental conditions, adding a coverslip above the sample is detrimental to the imaging.
5. Transfer to the microscope
- If the microscope is in a different room, prepare an insulated box with warmed gel packs to transfer the tissue sections while maintaining the temperature. The box also serves to keep the sections warm until they are imaged.
- Put the sample under the microscope, and make sure it is positioned properly under the objective by direct observation through the oculars with transmitted light.
NOTE: The goal is to align the predicted emitting structures with the oscillation direction of the laser to maximize the emitted (and, therefore, detected) SHG signal. - Remove the excess HBSS so that a thin liquid film covers the entire sample. Visually check the liquid film every few minutes to avoid excessive evaporation and drying of the sample.
NOTE: In the conditions described, a section is used for no more than 20 min. Drying of the sample causes drastic artifactual effects (Supplementary Figure 1A). Rather than adding more HBSS, periodic soaking of the section in warm, fresh medium is recommended if longer imaging times are needed. The use of perfusion chambers and glue or low-melting agarose to immobilize the tissue is also recommended when longer experiments are to be done. - Prepare the microscope stage for non-descanned imaging, which might include closing all the doors of the dark incubation chamber or covering the incubation chamber with a black nylon polyurethane-coated fabric.
6. Imaging
- Select the "non-descanned" imaging mode along the transmission path. This way, the capture of the weak SH signal of tubulin will be optimized.
- Select the LCI Plan-Neofluar 25x/0.8 NA objective.
- Set a laser power between 13 mW and 26 mW, with a pixel dwell time of 12.6 µs. Take images no bigger than 512 pixels x 512 pixels, with speed 5 and averaging 2, for an average acquisition time of about 15 s.
NOTE: Using higher laser powers and/or longer dwell times may damage the sample (Supplementary Figure 1B). - Take images first using a 485 nm short-pass filter (SP485), and, in a second step, add a sharp 405 nm bandpass filter (BP405; Figure 3). Follow the steps reported in Supplementary File 2.
NOTE: Pseudo-brightfield images can be taken through the same detector using the residual laser light of a visible line (405 nm or 488 nm lasers work best).
7. Processing of the cerebellum
- First, cut the cerebellum with a scalpel into two hemispheres, and then glue them to the vibratome support by the middle portion (the flat part obtained after cutting).
- Section and image the cerebellum in the same way as was described for the brain (160 µm sections, same vibratome settings, same blade, same microscope settings, etc.).
NOTE: Due to the anatomy of the cerebellum, its orientation at the moment of sectioning is not critical; in most of the slices generated away from the surface, there will be folia sectioned well into the white matter.
Representative Results
The images obtained with this methodology have an intrinsic low background level due to the very limited number of harmonophores present in biological tissues, which is one of the significant advantages of the method.
When the fibers of the corpus callosum are imaged, fiber-like short structures and rounded elements can be consistently found in the taiep brain (Figure 3B), while the corpus callosum of the control brain shows a much more heterogeneous and isotropic signal throughout the brain region (Figure 3A). The origin of the differential signal lies specifically in the second harmonic generation phenomenon, since adding the narrow bandpass filter only decreases the non-specific signal intensity from the control images (Figure 3C–D) while selectively removing this low, diffuse signal from around the soma-like and the short, elongated structures in the taiep images, which always generate intense SH light (Figure 3E–F).
The other structure analyzed, the cerebellar white matter, gives comparable results. Specifically, while in control tissue there is an almost complete absence of SH signal (Figure 4B), and the Purkinje cells are barely visible when using the short-pass filter, the elongated and rounded structures persist in the SH image from the taiep tissue (Figure 4D).
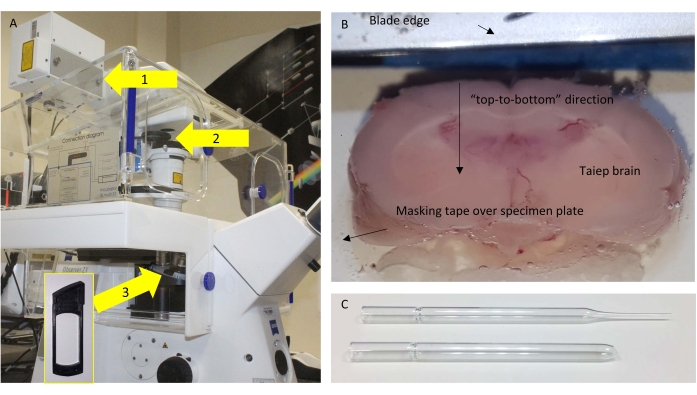
Figure 1: Microscope and sectioning. (A) Schematic of the microscope used, with relevant components highlighted. Arrows: 1 = NDD port where the SP485 is located close to the detector; 2 = removable frames where the BP405 is placed; 3 = position under the objective where the half-wave plate (HWP) is placed for control experiments. The inset shows the frame where the HWP could be inserted. (B) Top view of the vibratome buffer tray with the glued brain ready to be fine-sectioned. (C) The original (top) and modified (bottom) Pasteur pipette used to transfer the sections. Please click here to view a larger version of this figure.
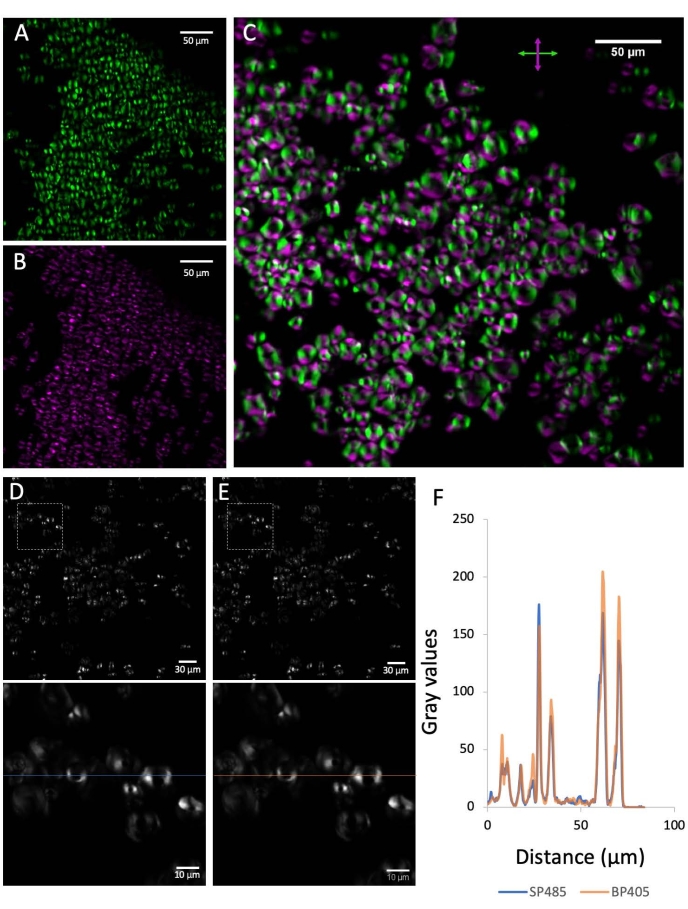
Figure 2: Preliminary controls. (A) The SHG signal emitted by corn starch grains shows a predominant orientation that coincides with the oscillation direction of the laser (horizontal in this case). (B) After inserting a half-wave plate with its fast axis oriented at 45° with respect to the horizontal, the signal is rotated by 90°. (C) Merging of the signal before and after the half-wave plate insertion. (D) The signal below 485 nm emitted from the corn starch grains. (E) The SHG signal from the starch grains detected at 405/10 nm. (F) Graph showing a comparison of the two signals corresponding to the line-scans shown in the zoomed inserts of D and E. The SHG filter used causes negligible signal loss. Please click here to view a larger version of this figure.
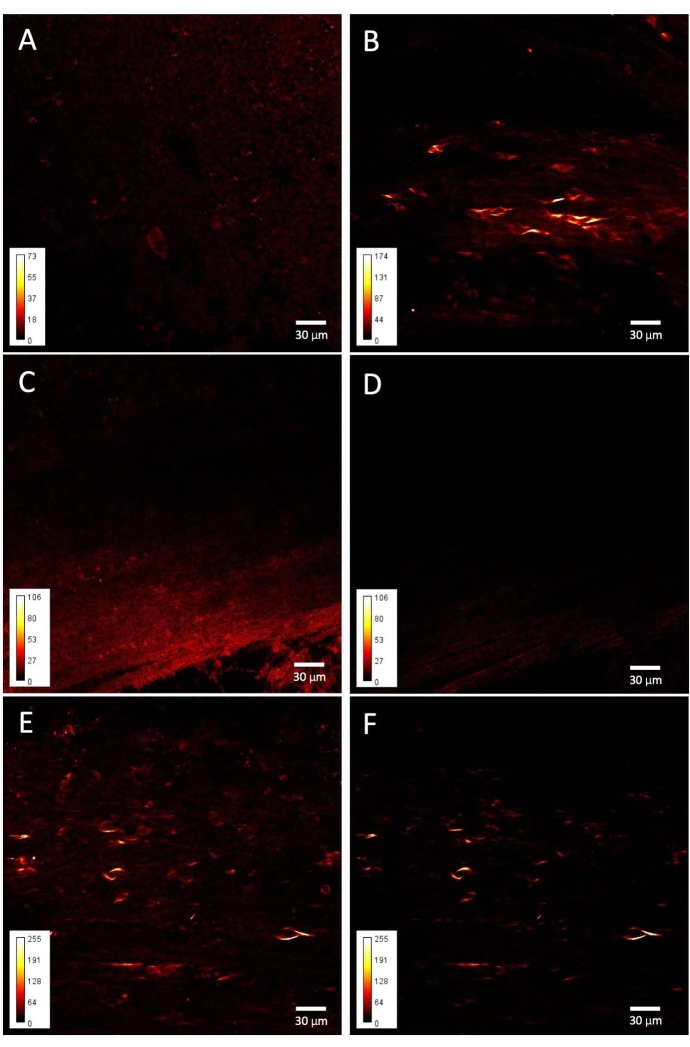
Figure 3: SHG images from taiep and WT corpus callosum. Representative examples of (A) WT and (B) taiep signals obtained from the corpus callosum. (C) SP485 filtered image from an WT corpus callosum. (D) BP405 filtered image from the same sample as in C. (E) SP485 filtered image from a taiep corpus callosum. (F) BP405 filtered image of the same sample as in E. Please click here to view a larger version of this figure.
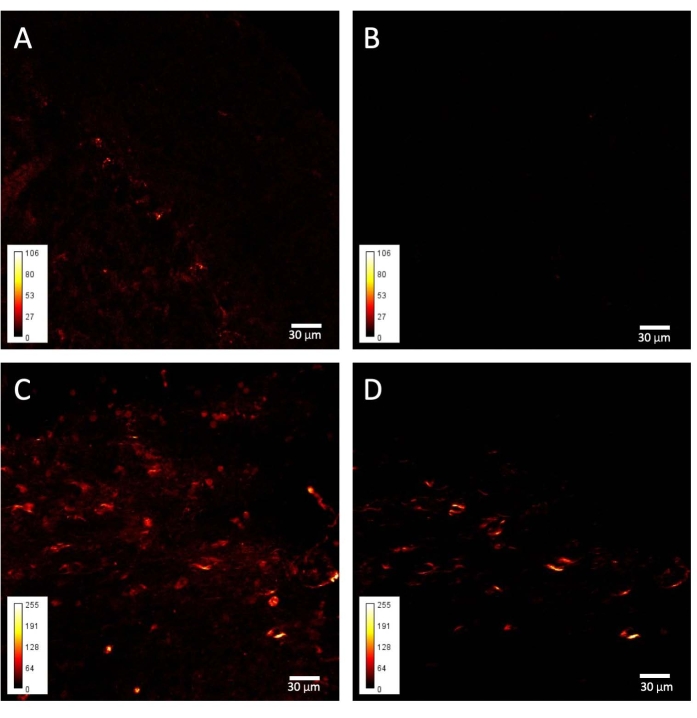
Figure 4: SHG images from taiep and WT cerebellums. Representative examples of (A–B) WT and (C–D) taiep signals obtained from cerebellar folia. (A) An SP485 filtered image from a WT folium; only some Purkinje cells are visible. (B) A BP405 filtered image from the same sample as in A. (C) An SP485 filtered image from a taiep folium. (D) A BP405 filtered image of the same sample as in C. Please click here to view a larger version of this figure.
Supplementary Figure 1: Examples of artifacts. (A) Artifact due to drying of the sample. (B) Artifact due to excessive exposure. Scale bars: 30 µm. Please click here to download this File.
Supplementary File 1: Köhler alignment.The file presents the steps for performing the Köhler alignment. Please click here to download this File.
Supplementary File 2: ZEN software steps for SHG image acquisition. Please click here to download this File.
Discussion
SHG microscopy is part of a group of non-linear optics techniques, which include two-photon excitation microscopy, third harmonic generation microscopy, and coherent anti-Stokes Raman scattering microscopy, that have contributed to expanding the range of applications of conventional optical microscopy to the life sciences20.
Specifically, the major strength and weakness of SHG microscopy relate to the same condition: the signal generator is non-centrosymmetric21. Such a specific architectural condition is often found in the realm of inorganic and organic crystals but is rare among biological objects. Together with collagen and muscular myosin, microtubules are able to generate a second harmonic signal22, which can be detected with a microscope if enough summation occurs in the bundles of parallel polymers. Inside the cells, tubulin associates to form parallel bundles in the cores of axons (a cell type-specific location), in mitotic spindles (a temporal-specific distribution), and, as presented in this work, in the trademark microtubule association of a recently described tubulinopathy-hypomyelination with atrophy of the basal ganglia and cerebellum, or H-ABC (which represents the first reported pathological distribution of this kind)14,16,23.
This sensory-motor syndrome is still an orphan disease, as many neurologists are not yet familiar with all the symptoms and/or cannot afford the genetic screening of suspected patients to confirm the diagnosis, which very likely causes underdiagnosis, at least in some populations. On top of that, not much is known about the pathology at the molecular and cellular level, so the techniques that could help shed light on the specific mechanisms of this degenerative process are very valuable, also at a basic level.
Therefore, two uses of SHG microscopy concerning this myelin disorder are proposed. In the long term, SHG microscopy could be integrated into diagnostic approaches for biopsy screening or direct intracranial analysis, but the greatest impact could be associated with trying to decipher the molecular basis of the disease.
There are many advantages of SHG microscopy over other microscopy techniques. Indeed, SHG microscopy does not require the fixation of the tissue, which is a particularly delicate step of sample preparation in the case of microtubules, and does not require staining of any kind. The greatest advantage is related to the physical phenomenon involved. On one hand, due to the virtually absent background, it provides high contrast despite the weak signals generated, and on the other hand, since the light intensities needed for frequency-doubling are very high and these intensities are met only in a very restricted volume of the sample, the technique provides intrinsic optical sectioning, thus allowing 3D reconstructions.
The major limitations of this microscopy technique are related to the cost of the equipment. A good confocal microscope, together with a pulsed infrared (IR) laser, is necessary for this application, and although a two-photon excitation system is available in many neuroscience labs, which can be easily modified into an SHG setup, the difference in cost compared to a conventional epifluorescence setup is still significant. Additionally, as a diffraction-limited technique, its resolution is affected by the use of IR light for illumination.
As with many pioneering applications, the setups used for the early experiments on SHG microscopy applied to the life sciences were totally customized optical setups built as open systems11,24,25, giving the experimenters the possibility to optimize every single component. Since, in life science labs, it is more common to have access to a commercial setup, we wanted to test whether one of these systems is sensitive enough to detect the weak signal emitted by tissular microtubules bundles, which are known to not be as strong harmonophores as collagen. The LSM710 NLO system by Zeiss consists of an inverted confocal microscope with modules for non-descanned detection in transmission and "epi" mode and with a Coherent Chameleon tunable IR laser. Due to the coherent nature of the SHG phenomenon, it is crucial to select the transmission path to maximize the collection of frequency-doubled light from the sample, and, therefore, the detection was performed through the 0.55 NA condenser by integration. An LCI Plan-Neofluar 25X/0.8 NA immersion objective was used to illuminate the sample from the bottom and approximate the condenser NA. With the conditions described in this paper, we could reliably detect a signal from the tissue bearing the TUBB4A mutation, and this signal was always absent in the controls.
In the reported protocol, two steps required adjustments to achieve the best imaging conditions: the thickness of the tissue section and the opening of the condenser diaphragm. The sections should be as thin as possible to prevent excessive absorbance, scattering, and loss of SH photons, especially since the goal is to image in transmission due to the coherent nature of the physical phenomenon. In our case, the limiting factor was the consistency of the taiep tissue, which hindered sectioning in homogeneously thick sections below 160-180 µm. With the conventional Köhler alignment, the condenser diaphragm is usually shut to about 60% of its area to generate contrast in the transmitted light image; conversely, for this application, the aim is to collect as many photons as possible, hence the total opening of the diaphragm.
This application of SHG microscopy is important as it provides the possibility for discerning tubulin-rich versus tubulin-normal tissue, with the tubulin-rich tissue emitting a detectable signal. It is also possible that other yet-to-be-described tubulinopathy mutations cause cytoskeletal disorders in other tissues26, which may also involve tubulin enrichment.
In terms of other possible applications, SHG microscopy could be expanded to the study of weaker signals, like those present in tubulin-normal tissue, in fine, dissociated axons, or in intracellular bundles with mixed polarity. To do so, setup modifications, such as the substitution of the air condenser with one with a higher numerical aperture and/or with immersion or the use of a more sensitive detector, could make an important difference.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACYT) through the following grants: infraestructura 226450 to VP-CIO, infraestructura 255277 to V.P., and FORDECYT-PRONACES/194171/2020 to V.H. We acknowledge the support of Juvenal Hernández Guevara at CIO in the video-making.
Materials
| 405/10 nm BrightLine(R) single-band bandpass filter | Semrock | FF01-405/10-25 | 32 mm diameter, with housing ring |
| Black Nylon, Polyurethane-Coated Fabric | Thorlabs | BK5 | 5' x 9' (1.5 m x 2.7 m) x 0.005" (0.12 mm) Thick |
| Blades for vibratome | any commercial; e.g. Wilkinson Sword | Classic stainless steel double edge razor blades | |
| Cell culture dishes, 35 mm | any commercial; e.g. Falcon | 351008 | |
| Confocal microscope | Zeiss | LSM710NLO AxioObserver Z1 | Inverted microscope, objective used is LCI Plan-Neofluar 25x/0.8 NA |
| Cooler | any commercial | Any insulated, polystyrene box could work, to mantain the sample at about 37 °C | |
| Corn stach | e.g. Maizena | From the supermarket | |
| Coverslips #1.5 | any commercial | Rectangular | |
| Cyanoacrylate glue | e.g. Loctite | To glue the brain to the masking tape | |
| Fine forceps | fine science tools | 11412-11 | To manipulate tissue sections by handling from the meninges |
| Fine scissors | fine science tools | 14370-22 | To cut the skin |
| Fine scissors curved tip | fine science tools | 14061-09 | To cut along the midline |
| Formaldehyde 37% | Sigma-Aldrich | 252549 | To dilute 1:10 in PBS |
| Friedman Rongeur | fine science tools | 16000-14 | To cut the bone |
| Gel packs | any commercial | Prewarmed to 37 °C, to help mantaining the temperature inside the cooler | |
| Glass Pasteur pipette, modified | any commercial | To transfer the tissue section | |
| Hanks′ Balanced Salt solution (HBSS) | Gibco | 14025-076 | Could be prepared from powders |
| Kelly hemostats | fine science tools | 13018-14 | To separate the bone |
| Masking tape | any commercial | To protect th surface of the specimen plate | |
| NDD module, type C | Zeiss | 000000-1410-101 | To detect the signal, reducing light loss. Housing the 000000-1935-163 filter set with the SP485 |
| Offset bone nippers | fine science tools | 16101-10 | To cut the bone |
| Phosphate buffered saline (PBS) | Gibco | 10010-031 | Could be prepared from powders or tabs |
| Pulsed laser | Coherent | Chameleon Vision II | 680–1080 nm tunable laser |
| Scalpel | any commercial | Straight blade with sharp point | |
| Standard pattern forceps | fine science tools | 11000-18 | |
| Vannas spring scissors | fine science tools | 15018-10 | To cut meninges that remain joined to both the slice obtained from vibratome cutting and the section glued to the specimen plate. |
| Vibratome | any commercial; e.g. Leica | VT1200 |
Referencias
- Palmiter, R. D., et al. Cell lineage ablation in transgenic mice by cell-specific expression of a toxin gene. Cell. 50 (3), 435-443 (1987).
- Banerjee, A., et al. A monoclonal antibody against the type II isotype of beta-tubulin. Preparation of isotypically altered tubulin. The Journal of Biological Chemistry. 263 (6), 3029-3034 (1988).
- Banerjee, A., Roach, M. C., Trcka, P., Luduena, R. F. Preparation of a monoclonal antibody specific for the class IV isotype of beta-tubulin. Purification and assembly of alpha beta II, alpha beta III, and alpha beta IV tubulin dimers from bovine brain. The Journal of Biological Chemistry. 267 (8), 5625-5630 (1992).
- Banerjee, A., et al. Localization of βv tubulin in the cochlea and cultured cells with a novel monoclonal antibody. Cell Motility and the Cytoskeleton. 65 (6), 505-514 (2008).
- Cross, A. R., Williams, R. C. Kinky microtubules: Bending and breaking induced by fixation in vitro with glutaraldehyde and formaldehyde. Cell Motility and the Cytoskeleton. 20 (4), 272-278 (1991).
- Van Steenbergen, V., et al. Molecular understanding of label-free second harmonic imaging of microtubules. Nature Communications. 10 (1), 3530 (2019).
- Campagnola, P. J., Clark, H. A., Mohler, W. A., Lewis, A., Loew, L. M. Second-harmonic imaging microscopy of living cells. Journal of Biomedical Optics. 6 (3), 277 (2001).
- Campagnola, P. J., et al. Three-dimensional high-resolution second-harmonic generation imaging of endogenous structural proteins in biological tissues. Biophysical Journal. 82 (1), 493-508 (2002).
- Yu, C. -. H., et al. Measuring microtubule polarity in spindles with second-harmonic generation. Biophysical Journal. 106 (8), 1578-1587 (2014).
- Bancelin, S., et al. Probing microtubules polarity in mitotic spindles in situ using Interferometric Second Harmonic Generation Microscopy. Scientific Reports. 7, 6758 (2017).
- Dombeck, D. A., et al. Uniform polarity microtubule assemblies imaged in native brain tissue by second-harmonic generation microscopy. Proceedings of the National Academy of Sciences of the United States of America. 100 (12), 7081-7086 (2003).
- Psilodimitrakopoulos, S., et al. Estimation of the effective orientation of the SHG source in primary cortical neurons. Optics Express. 17 (16), 14418 (2009).
- Sharoukhov, D., Bucinca-Cupallari, F., Lim, H. Microtubule imaging reveals cytoskeletal deficit predisposing the retinal ganglion cell axons to atrophy in DBA/2J. Investigative Opthalmology & Visual Science. 59 (13), 5292 (2018).
- Alata, M., Piazza, V., Eguibar, J. R., Cortes, C., Hernandez, V. H. H-ABC tubulinopathy revealed by label-free second harmonic generation microscopy. Scientific Reports. 12, 14417 (2022).
- Duncan, I. D., Lunn, K. F., Holmgren, B., Urba-Holmgren, R., Brignolo-Holmes, L. The taiep rat: A myelin mutant with an associated oligodendrocyte microtubular defect. Journal of Neurocytology. 21 (12), 870-884 (1992).
- Duncan, I. D., et al. A mutation in the Tubb4a gene leads to microtubule accumulation with hypomyelination and demyelination: Tubb4a Mutation. Annals of Neurology. 81 (5), 690-702 (2017).
- Garduno-Robles, A., et al. MRI features in a rat model of H-ABC tubulinopathy. Frontiers in Neuroscience. 14, 555 (2020).
- Lopez-Juarez, A., et al. Auditory impairment in H-ABC tubulinopathy. Journal of Comparative Neurology. 529 (5), 957-968 (2021).
- Alata, M., et al. Longitudinal evaluation of cerebellar signs of H-ABC tubulinopathy in a patient and in the taiep model. Frontiers in Neurology. 12, 702039 (2021).
- Parodi, V., et al. Nonlinear optical microscopy: From fundamentals to applications in live bioimaging. Frontiers in Bioengineering and Biotechnology. 8, 585363 (2020).
- Lefort, C. A review of biomedical multiphoton microscopy and its laser sources. Journal of Physics D: Applied Physics. 50 (42), 423001 (2017).
- Campagnola, P. J., Loew, L. M. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nature Biotechnology. 21 (11), 1356-1360 (2003).
- vander Knaap, M. S., et al. New syndrome characterized by hypomyelination with atrophy of the basal ganglia and cerebellum. American Journal of Neuroradiology. 23 (9), 1466 (2002).
- Stoller, P., Kim, B. -. M., Rubenchik, A. M., Reiser, K. M., Da Silva, L. B. Polarization-dependent optical second-harmonic imaging of a rat-tail tendon. Journal of Biomedical Optics. 7 (2), 205 (2002).
- Brown, E. B., et al. In vivo measurement of gene expression, angiogenesis and physiological function in tumors using multiphoton laser scanning microscopy. Nature Medicine. 7 (7), 864-868 (2001).
- Chakraborti, S., Natarajan, K., Curiel, J., Janke, C., Liu, J. The emerging role of the tubulin code: From the tubulin molecule to neuronal function and disease. Cytoskeleton. 73 (10), 521-550 (2016).

