Absolute Quantification of Cell-Free Protein Synthesis Metabolism by Reversed-Phase Liquid Chromatography-Mass Spectrometry
Summary
Here, we present a robust protocol to quantify 40 compounds involved in central carbon and energy metabolism in cell-free protein synthesis reactions. The cell-free synthesis mixture is derivatized with aniline for effective separation using reversed-phase liquid chromatography and then quantified by mass spectrometry using isotopically labelled internal standards.
Abstract
Cell-free protein synthesis (CFPS) is an emerging technology in systems and synthetic biology for the in vitro production of proteins. However, if CFPS is going to move beyond the laboratory and become a widespread and standard just in time manufacturing technology, we must understand the performance limits of these systems. Toward this question, we developed a robust protocol to quantify 40 compounds involved in glycolysis, the pentose phosphate pathway, the tricarboxylic acid cycle, energy metabolism and cofactor regeneration in CFPS reactions. The method uses internal standards tagged with 13C-aniline, while compounds in the sample are derivatized with 12C-aniline. The internal standards and sample were mixed and analyzed by reversed-phase liquid chromatography-mass spectrometry (LC/MS). The co-elution of compounds eliminated ion suppression, allowing the accurate quantification of metabolite concentrations over 2-3 orders of magnitude where the average correlation coefficient was 0.988. Five of the forty compounds were untagged with aniline, however, they were still detected in the CFPS sample and quantified with a standard curve method. The chromatographic run takes approximately 10 min to complete. Taken together, we developed a fast, robust method to separate and accurately quantify 40 compounds involved in CFPS in a single LC/MS run. The method is a comprehensive and accurate approach to characterize cell-free metabolism, so that ultimately, we can understand and improve the yield, productivity and energy efficiency of cell-free systems.
Introduction
Cell-free protein synthesis (CFPS) is a promising platform for manufacturing of proteins and chemicals, an application that has traditionally been reserved for living cells. Cell-free systems are derived from crude cell extracts and eliminate the complications associated with cell growth1. In addition, CFPS allows for direct access to metabolites and the biosynthetic machinery without the interference of a cell wall. However, a fundamental understanding of the performance limits of cell-free processes has been lacking. High-throughput methods for metabolite quantification are valuable for the characterization of metabolism and are critical for the construction of metabolic computational models2,3,4. Common methods used to determine metabolite concentrations include nuclear magnetic resonance (NMR), Fourier transform-infrared spectroscopy (FT-IR), enzyme-based assays, and mass spectrometry (MS)5,6,7,8. However, these methods are often limited by their inability to efficiently measure multiple compounds at once and often require a sample size greater than typical cell-free reactions. For example, enzyme-based assays can often only be used to quantify a single compound in a run, and are limited when the sample size is small, such as in cell-free protein synthesis reactions (typically run on a 10-15 μL scale). Meanwhile, NMR requires a high abundance of metabolites for detection and quantification5. Toward these shortcomings, chromatography methods in tandem with mass spectrometry (LC/MS) provide several advantages, including high sensitivity and the capability of measuring multiple species simultaneously9; however, the analytical complexity increases considerably with the number and diversity of species being measured. It is important, therefore, to develop methods that fully realize the high-throughput potential of LC/MS systems. Compounds in a sample are separated by liquid chromatography and identified through mass spectrometry. The signal of the compound depends on its concentration and ionization efficiency, where the ionization can vary between compounds and may also depend on the sample matrix.
Achieving the same ionization efficiency between the sample and standards is a challenge when using LC/MS to quantify analytes. Further, quantification becomes more challenging with metabolite diversity due to signal splitting and heterogeneity in proton affinity and polarity10. Lastly, the co-eluting matrix of the sample can also affect the ionization efficiencies of the compounds. To address these issues, metabolites can be chemically derivatized, increasing the separation resolution and sensitivity by LC/MS systems, while simultaneously decreasing signal splitting in some cases10,11. Chemical derivatization works by tagging specific functional groups of metabolites to adjust their physical properties like charge or hydrophobicity to increase ionization efficiency11. Various tagging agents can be used to target different functional groups (e.g., amines, hydroxyls, phosphates, carboxylic acids, etc.). Aniline, one such derivatization agent, targets multiple functional groups at once, and adds a hydrophobic component into hydrophilic molecules, thereby increasing their separation resolution and signal12. To address the co-eluting matrix ion suppression effect, Yang and coworkers developed a technique based on Group Specific Internal Standard Technology (GSIST) labeling where standards are tagged with 13C aniline isotopes and mixed with the sample12,13. The metabolite and corresponding internal standard have the same ionization efficiency since they co-elute, and their intensity ratio can be used to quantify the concentration in the experimental sample.
In this study, we developed a protocol to detect and quantify 40 compounds involved in glycolysis, the pentose phosphate pathway, the tricarboxylic acid cycle, energy metabolism and cofactor regeneration in CFPS reactions. The method is based on the GSIST approach, where we used 12C-aniline and 13C-aniline to tag, detect, and quantify metabolites using reversed-phase LC/MS. The linear range of all compounds spanned 2-3 orders of magnitude with an average correlation coefficient of 0.988. Thus, the method is a robust and accurate approach to interrogate cell-free metabolism, and possibly whole-cell extracts.
Protocol
1. Preparation of reagents for aniline tagging
- Prepare a 6 M aniline solution at pH 4.5. Working in a hood, combine 550 µL of aniline with 337.5 µL of LCMS grade water and 112.5 µL of 12 M hydrochloric acid (HCl) in a centrifuge tube. Vortex well and store at 4 °C.
NOTE: Aniline can be stored at 4 °C for 2 months.
CAUTION: Aniline is highly toxic and should be worked with in a fume hood. Hydrochloric acid is highly corrosive - Prepare a 6 M 13C aniline solution at pH 4.5. Combine 250 mg of 13C6-aniline with 132 µL of water and 44 µL of 12 M HCl. Vortex well and store at 4 °C.
- Prepare 200 mg/mL N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC) solution. Dissolve 2 mg of EDC in 10 µL of water for every sample to be tagged and vortex well.
NOTE: EDC solution should be prepared the same day as the reaction. EDC acts as a catalyst for the derivatization of compounds with aniline12.
2. Preparation of standards
- Make separate stock solutions of all compounds dissolved in LC/MS grade water (Table 1).
- Preparation of internal standard stock solution
- Combine all compounds except for nicotinamide adenine dinucleotide (NAD), nicotinamide adenine dinucleotide phosphate (NADP), flavin adenine dinucleotide (FAD), acetyl coenzyme A (ACA), and glycerol 3-phosphate (Gly3P), with the appropriate volumes to create a 2 mM stock solution of all compounds.
- Combine NAD, NADP, FAD, ACA, and Gly3P with the appropriate volumes to create a 2 mM stock solution.
3. Preparation of sample (Figure 1)
- Quench and precipitate the proteins in a cell-free protein synthesis reaction by adding an equal volume of ice-cold 100% ethanol to the reaction. Centrifuge the sample at 12,000 x g for 15 min at 4 °C. Transfer the supernatant to a new centrifuge tube.
NOTE: Samples can be stored at -80 °C at this point and analyzed at a later time
4. Labeling reaction
- Labeling sample with 12C-aniline solution
- Transfer 6 µL of sample into a new centrifuge tube and bring the volume to 50 µL with water.
NOTE: Volume sample size may depend on the specific CFPS reaction. - Add 5 µL of 200 mg/mL EDC solution.
- Add 5 µL of 12C-aniline solution.
NOTE: The aniline solution separates into two phases. Mix well before adding to the reaction. - Vortex the reaction with gentle shaking for 2 h at room temperature.
- After 2 h, remove the tubes from the shaker and add 1.5 µL of triethylamine (TEA) to the reaction in a fume hood.
NOTE: Triethylamine raises the pH of the solution which stops the aniline tagging reaction and stabilizes the compounds.
CAUTION: Triethylamine is toxic and causes irritation of the eyes and respiratory tract. - Centrifuge at 13,500 x g for 3 min.
- Transfer 6 µL of sample into a new centrifuge tube and bring the volume to 50 µL with water.
- Labeling internal standards with 13C-aniline solution
- Dilute internal stock solution to 80 µM with a final volume of 50 µL.
NOTE: Concentration of internal standards can be adjusted to levels close to the experimental sample. - Add 5 µL of 200 mg/mL EDC solution.
- Add 5 µL of 13C-aniline solution.
- Vortex the reaction with gentle shaking for 2 h at room temperature.
- After 2 h, remove the tubes from the shaker and add 1.5 µL of TEA to the reaction in a fume hood.
- Centrifuge at 13,500 x g for 3 min.
- Dilute internal stock solution to 80 µM with a final volume of 50 µL.
- Combining tagged internal standard and tagged sample
- Mix 25 µL of 12C-aniline labeled sample with 25 µL of 13C-aniline labeled standard.
- Transfer to an auto-sampler vial and analyze by the LC/MS procedure.
- Creating a standard curve for untagged metabolites
- Dilute stock solution of untagged metabolites (NAD, NADP, FAD, ACA, and Gly3P) to final concentrations of 320 µM, 80 µM, 20 µM and 5 µM with a volume of 50 µL.
- Add 5 µL of 200 mg/mL EDC solution.
- Add 5 µL of 12C-aniline solution.
- Vortex the reaction with gentle shaking for 2 h at room temperature.
- After 2 h, remove the tubes from the shaker and add 1.5 µL of TEA to the reaction in a fume hood.
- Centrifuge at 13,500 x g for 3 min.
- Transfer supernatant to an auto-sampler vial and analyze by the LC/MS procedure.
NOTE: The untagged metabolites follow the same procedure as the sample to replicate the sample matrix in order to maintain similar ionization efficiency.
5. Setup of LC/MS procedure
- Preparation of solvents
- Prepare 5 mM tri-butylamine (TBA) aqueous solution adjusted to pH 4.75 with acetic acid.
NOTE: TBA in the mobile phase helps the analytes achieve good resolution and separation14. - Prepare 5 mM TBA in acetonitrile (ACN).
- Prepare wash solvent with 5% water and 95% ACN.
- Prepare purge solvent with 95% water and 5% ACN.
- Prepare 5 mM tri-butylamine (TBA) aqueous solution adjusted to pH 4.75 with acetic acid.
- Setup of MS conditions
- Set the mass spectrometer to negative ion mode with a probe temperature of 520 °C, negative capillary voltage of -0.8 kV, positive capillary voltage of 0.8 kV, and set the software to acquire data at 5 points/s.
- Set selected ion recordings (SIR) for each metabolite with specified cone voltages and mass over charge (m/z) values. See Table 1.
- Initializing LC/MS according to manufacturer’s instructions
- Prime solvent lines in the solvent manager for 3 min.
- Prime wash solvent (5% water, 95% ACN) and purge solvent (95% water, 5% ACN) for 15 s for 5 cycles.
- Set the sample manager to 10 °C.
- Install a C18 (1.7µm, 2.1mm x 150mm) column and initialize column with 100% ACN at 0.3 mL/min for 10 min.
- Condition the column at 95% water and 5% ACN at 0.3 mL/min for 10 min prior to introducing solvents with buffers.
- Condition the column at 95% solvent A (5mM TBA aqueous, pH 4.75) and 5% solvent B (5 mM TBA in ACN) at 0.3 mL/min for 10 min.
- Set up a gradient protocol with the elution starting at 95% solvent A and 5% solvent B, raised to 70% solvent B in 10 min, raised to 100% solvent B in 2 min and held at 100% solvent B for 3 min. Return to initial conditions (95% solvent A, 5% solvent B) over 1 min and hold for 9 min to re-equilibrate the column.
- Condition the column with the gradient protocol 3 times prior to any injections onto the column.
- Injecting sample and standards
- Inject 5 µL of the sample into the column and acquire the appropriate m/z ion intensities for the 12C-aniline tagged sample.
- Inject 5 µL of the same sample again, but this time acquire the m/z ion intensities for the 13C-aniline tagged standards.
NOTE: Our LC/MS system is unable to acquire both 12C and 13C m/z intensities at the specified SIR time windows, since it is too much data to acquire in the specified time window. Therefore, we inject the same sample twice. - Inject untagged metabolite standards from lowest concentration to highest and record the appropriate m/z ion intensities.
6. Quantification
- Creating Export method
- In data acquisition software, select File > New Method > Export Method.
- Specify a Filename, such as AnilineTagging_Date.
- Check the Export ASCII File and choose a directory to export the text file to.
- In Report Type, select Summary by All.
- In Delimiters, for Column select a ,. For Row, select [cr][if].
- In Table, select Export and then Edit Table to include SampleName, Area, Alto, Amount and Units.
- Save export method.
- Quantifying metabolites with internal standards using data acquisition software
- Under the Sample Sets tab, right click the corresponding LC/MS run and select View as > Channels.
- Select all SIR channels for the 13C-aniline internal standards of one injection, right click and select Review.
- If the LC Processing Method Layout window does not automatically appear, go to View > Processing Method Layout.
- In Processing Method Layout, go to the Integration tab and set ApexTrack as the algorithm.
- Go to the Smoothing tab and set the type to Mean and the smoothing level to 13.
NOTE: Any smoothing level can be selected, as long as it is consistent across all samples. - In the MS Channel tab, disable MS 3D Processing.
- In the SIR channel window, integrate each peak, one channel at a time. Once a peak is integrated, go to Options > Fill from Result and the details of the peak will be filled in the Components tab. Change the peak name to the corresponding compound name.
- Once all the SIR channels have been evaluated, save the processing method and close window.
- Select all SIR channels of the 13C-aniline and 12C-aniline tagged sample, right click and select Process.
- Check the Process box, select Use specified processing method, and choose the processing method that is just saved. Also check the Export box, select Use specified export method and choose the saved export method created earlier. Click OK.
- Open the exported text file with Excel and calculate the concentration of the unknown compound using:
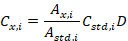
where Cx,i is the concentration of the unknown sample for metabolite i, Ax,i is the integrated area of the unknown metabolite i, Astd,i is the integrated area of the internal standard of metabolite i, Cstd,i is the concentration of the internal standard of metabolite i, and D is the dilution factor.
- Quantifying untagged metabolites with standard curve
- Under the Sample Sets tab, right click the corresponding LC/MS run and select View as > Channels.
- Select all SIR channels for the untagged standards of one injection, right click and select Review.
- If the LC Processing Method Layout window does not automatically appear, go to View > Processing Method Layout.
- In Processing Method Layout, go to the Integration tab and set ApexTrack as the algorithm.
- Go to the Smoothing tab and set the type to Mean and the smoothing level to 13.
NOTE: Any smoothing level can be selected, as long as it is consistent across all samples. - In the MS Channel tab, disable MS 3D Processing.
- In the SIR channel window, integrate each peak, one channel at a time. Once a peak is integrated, go to Options > Fill from Result and the details of the peak will be filled in the Components tab. Change the peak name to the corresponding compound name.
- Once all the SIR channels have been evaluated, save the processing method and close window.
- Under the Sample Sets tab, right click on the sample set and select Alter Sample.
- Select Amount in the new window.
- Select copy from Process method and choose the process method that was just saved.
- Enter the concentration of each metabolite for each vial and enter the unit as <μM for each component (or the corresponding unit) and select OK.
- Select the sample set again, right click, View as > Channels.
- Select all SIR channels of the untagged metabolites for the standards, right click and select Process.
- Check the Process box and choose Use specified processing method. Select the appropriate processing method and click OK.
- Select SIR channels for all untagged metabolites for the samples, right click and select Process.
- Check the Process box, select Use specified processing method, and choose the processing method that was just saved. Also check the Export box, select Use specified export method and choose the saved export method created earlier. Click OK.
- Quantify the untagged metabolites with the standard curve and export the results to a text file to the directory specified.
Representative Results
As a proof-of-concept, we used the protocol to quantify metabolites in an E. coli based CFPS system expressing green fluorescent protein (GFP). The CFPS reaction (14 μL) was quenched and deproteinized with ethanol. The CFPS sample was then tagged with 12C-aniline, while standards were tagged with 13C-aniline. The tagged sample and standards were then combined and injected into the LC/MS (Figure 1). The protocol detected and quantified 40 metabolites involved in central carbon and energy metabolism using internal standards, while a standard curve for 5 of the metabolites that were not tagged with aniline was also developed (Figure 2). The diverse metabolites involved in these pathways were a class of phosphorylated sugars, phosphocarboxylic acids, carboxylic acids, nucleotides, and cofactors. The derivatization with aniline introduced a hydrophobic moiety into hydrophilic molecules which facilitated more effective separation using reversed-phase chromatography12. In addition, the method enabled the separation of structural isomer pairs such as glucose 6-phosphate and fructose 6-phosphate in a single LC/MS run. Each compound’s mass over charge (m/z) ratio and retention time were identified prior to the experiment by injecting 1 mM of one compound at a time and comparing the mass spectrum to the blank (Table 1).
The limit of detection and range of linearity for all compounds was estimated by producing a standard curve that ranged from 0.10 µM to 400 µM (Table 2). The average correlation coefficient (R2) for all compounds was 0.988 and most compounds had a linear range of 3-orders of magnitude. Three compounds had notable saturation effects, especially alpha-ketoglutarate which had a linear range from 0.1 µM to 25 µM. Isocitrate and citrate also had saturation effects above 100 µM.
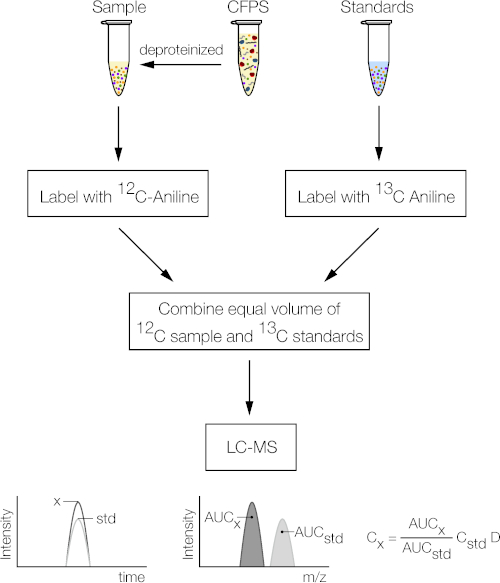
Figure 1: Schematic of workflow for aniline tagging. The cell-free protein synthesis reaction is deproteinized and tagged with 12C-aniline, while a standard stock mixture is tagged with 13C-aniline. Both mixtures are then mixed at a 1:1 volumetric ratio and analyzed by LC/MS. Please click here to view a larger version of this figure.
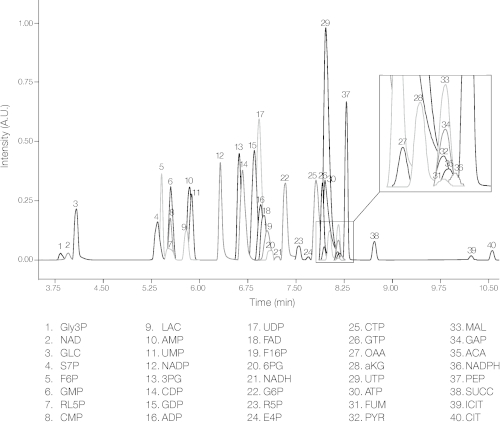
Figure 2: Overlapped selected ion chromatograms for 40 metabolites. Mass chromatogram from a single LC/MS run of a 40 µM standard mixture of 40 metabolites. Peaks were identified by their retention time and m/z values for each compound. Complete compound names and their abbreviations are listed in Table 1. Please click here to view a larger version of this figure.
| Peak Number | Metabolite | Abbreviation | KEGG ID | Retention Time (min) | 12C m/z | 13C m/z | nonlabel m/z | CV | MS Species | |
| 1 | Glycerol 3-phosphate | Gly3P | C00093 | 3.85 | 153 | 10 | M – H2O – H | |||
| 2 | Nicotinamide adenine dinucleotide | NAD | C00003 | 3.96 | 698 | 10 | M + Cl – H | |||
| 3 | Glucose | GLC | C00031 | 4.06 | 289.9 | 296 | 15 | M + A + Cl – H | ||
| 4 | Sedoheptulose 7-phosphate | S7P | C05382 | 5.41 | 364 | 370 | 10 | M + A – H | ||
| 5 | Fructose 6-phosphate | F6P | C00085 | 5.48 | 334 | 340 | 10 | M + A – H | ||
| 6 | Guanosine monophosphate | GMP | C00144 | 5.57 | 437.05 | 443 | 10 | M + A – H | ||
| 7 | Ribulose 5-phosphate | RL5P | C00199 | 5.58 | 304 | 310 | 10 | M + A – H | ||
| 8 | Cytidine monophosphate | CMP | C00055 | 5.59 | 397.09 | 403 | 10 | M + A – H | ||
| 9 | Lactate | LAC | C00186 | 5.77 | 164.05 | 170 | 10 | M + A – H | ||
| 10 | Adenosine monophosphate | AMP | C00020 | 5.85 | 421.1 | 427.1 | 10 | M + A – H | ||
| 11 | Uridine monophosphate | UMP | C00105 | 5.88 | 398.07 | 404 | 10 | M + A – H | ||
| 12 | Nicotinamide adenine dinucleotide phosphate | NADP | C00006 | 6.39 | 724 | 10 | M – H2O – H | |||
| 13 | 3-Phosphoglyceric acid | 3PG | C00197 | 6.63 | 242 | 248.06 | 15 | M + A – H2O – H | ||
| 14 | Cytidine diphosphate | CDP | C00112 | 6.72 | 477 | 483 | 10 | M + A – H | ||
| 15 | Guanosine diphosphate | GDP | C00035 | 6.87 | 517 | 523 | 10 | M + A – H | ||
| 16 | Adenosine diphosphate | ADP | C00008 | 6.94 | 501 | 507 | 10 | M + A – H | ||
| 17 | Uridine diphosphate | UDP | C00015 | 6.97 | 478 | 484 | 10 | M + A – H | ||
| 18 | Flavin adenine dinucleotide | FAD | C00016 | 7.03 | 784.15 | 15 | M – H | |||
| 19 | Fructose 1,6-bisphosphate | F16P | C05378 | 7.1 | 395.95 | 402.1 | 10 | M + A – H2O – H | ||
| 20 | Gluconate 6-phosphate | 6PG | C00345 | 7.11 | 425.1 | 437 | 10 | M + 2A – H | ||
| 21 | Nicotinamide adenine dinucleotide reduced | NADH | C00004 | 7.23 | 633.13 | 639.08 | 10 | M + A + H2O – nicotinamide – H | ||
| 22 | Glucose 6-phosphate | G6P | C00668 | 7.32 | 409.1 | 421.1 | 10 | M + 2A – H | ||
| 23 | Ribose 5-phosphate | R5P | C00117 | 7.54 | 379.1 | 391.1 | 15 | M + 2A – H | ||
| 24 | Erythrose 4-phosphate | E4P | C00279 | 7.71 | 348.9 | 361 | 10 | M + 2A – H | ||
| 25 | Cytidine triphosphate | CTP | C00075 | 7.84 | 557 | 563 | 5 | M + A – H | ||
| 26 | Guanosine triphosphate | GTP | C00044 | 7.93 | 597 | 603 | 5 | M + A – H | ||
| 27 | Oxalacetate | OAA | C00036 | 7.94 | 281 | 293 | 25 | M + 2A – H | ||
| 28 | Alpha-ketoglutarate | aKG | C00026 | 7.95 | 295 | 307.1 | 15 | M + 2A – H | ||
| 29 | Uridine triphosphate | UTP | C00075 | 7.97 | 558 | 564 | 10 | M + A – H | ||
| 30 | Adenosine triphosphate | ATP | C00002 | 8.03 | 581 | 587 | 15 | M + A – H | ||
| 31 | Fumarate | FUM | C00122 | 8.09 | 265 | 277.1 | 10 | M + 2A – H | ||
| 32 | Pyruvate | PYR | C00022 | 8.09 | 162 | 168 | 25 | M + A – H | ||
| 33 | Malate | MAL | C00149 | 8.09 | 283.06 | 295.15 | 10 | M + 2A – H | ||
| 34 | D-glyceraldehyde 3-phosphate | GAP | C00118 | 8.09 | 319 | 331.1 | 5 | M + 2A – H | ||
| 35 | Acetyl-coenzyme A | ACA | C00024 | 8.16 | 790 | 10 | M – H2O – H | |||
| 36 | Nicotinamide adenine dinucleotide phosphate reduced | NADPH | C00005 | 8.23 | 694.92 | 700.82 | 10 | M + A – nicotinamide – H | ||
| 37 | Phosphoenolpyruvate | PEP | C00074 | 8.28 | 317 | 329.1 | 20 | M + 2A – H | ||
| 38 | Succinate | SUCC | C00042 | 8.64 | 267.07 | 279.1 | 15 | M + 2A – H | ||
| 39 | Isocitrate | ICIT | C00311 | 10.13 | 398 | 416 | 10 | M + 3A – H2O – H | ||
| 40 | Citrate | CIT | C00158 | 10.46 | 416.1 | 434.06 | 20 | M + 3A – H | ||
Table 1: Identification and labeling results of metabolites. Each compound’s corresponding peak number, retention time, m/z value for unlabeled, 12C and 13C labeled, and MS species. MS Species, A stands for Aniline tag.
| Peak No. | Metabolite | Abbreviation | KEGG ID | Concentration (mM) | SD (n = 3) | Limit of Detection (μM) | Limit of Linear Range (μM) | R^2 | |
| 1 | Glycerol 3-phosphate | Gly3P | C00093 | 0.377 | 0.034 | 0.1 | 400 | 0.995 | |
| 2 | Nicotinamide adenine dinucleotide | NAD | C00003 | 0.052 | 0.010 | 0.39 | 400 | 0.993 | |
| 3 | Glucose | GLC | C00031 | 0.002 | 0.000 | 0.1 | 400 | 0.997 | |
| 4 | Sedoheptulose 7-phosphate | S7P | C05382 | 0.007 | 0.000 | 0.16 | 400 | 0.988 | |
| 5 | Fructose 6-phosphate | F6P | C00085 | 0.029 | 0.004 | 0.1 | 400 | 0.986 | |
| 6 | Guanosine monophosphate | GMP | C00144 | 0.007 | 0.001 | 0.39 | 100 | 0.992 | |
| 7 | Ribulose 5-phosphate | RL5P | C00199 | 0.035 | 0.002 | 0.39 | 400 | 0.996 | |
| 8 | Cytidine monophosphate | CMP | C00055 | 0.045 | 0.001 | 0.1 | 100 | 0.992 | |
| 9 | Lactate | LAC | C00186 | 2.134 | 0.048 | 0.1 | 400 | 0.988 | |
| 10 | Adenosine monophosphate | AMP | C00020 | 0.020 | 0.002 | 0.1 | 100 | 0.992 | |
| 11 | Uridine monophosphate | UMP | C00105 | 0.021 | 0.000 | 0.1 | 100 | 0.997 | |
| 12 | Nicotinamide adenine dinucleotide phosphate | NADP | C00006 | 0.014 | 0.002 | 0.34 | 400 | 0.950 | |
| 13 | 3-Phosphoglyceric acid | 3PG | C00197 | 6.125 | 0.239 | 0.1 | 100 | 0.996 | |
| 14 | Cytidine diphosphate | CDP | C00112 | 0.202 | 0.029 | 0.39 | 400 | 0.997 | |
| 15 | Guanosine diphosphate | GDP | C00035 | 0.146 | 0.027 | 1.5625 | 400 | 0.984 | |
| 16 | Adenosine diphosphate | ADP | C00008 | 0.797 | 0.161 | 0.39 | 400 | 0.995 | |
| 17 | Uridine diphosphate | UDP | C00015 | 0.212 | 0.036 | 0.39 | 400 | 0.991 | |
| 18 | Flavin adenine dinucleotide | FAD | C00016 | 0.008 | 0.001 | 0.1 | 400 | 0.958 | |
| 19 | Fructose 1,6-bisphosphate | F16P | C05378 | 3.643 | 0.105 | 0.39 | 400 | 0.989 | |
| 20 | Gluconate 6-phosphate | 6PG | C00345 | 0.017 | 0.001 | 0.39 | 400 | 0.989 | |
| 21 | Nicotinamide adenine dinucleotide reduced | NADH | C00004 | 0.063 | 0.028 | 0.39 | 100 | 0.972 | |
| 22 | Glucose 6-phosphate | G6P | C00668 | 0.046 | 0.002 | 0.1 | 400 | 0.984 | |
| 23 | Ribose 5-phosphate | R5P | C00117 | 0.055 | 0.005 | 0.39 | 100 | 0.999 | |
| 24 | Erythrose 4-phosphate | E4P | C00279 | 0.038 | 0.007 | 0.39 | 400 | 0.979 | |
| 25 | Cytidine triphosphate | CTP | C00075 | 0.896 | 0.078 | 6.25 | 100 | 0.998 | |
| 26 | Guanosine triphosphate | GTP | C00044 | 0.870 | 0.109 | 6.25 | 100 | 0.993 | |
| 27 | Oxalacetate | OAA | C00036 | 0.023 | 0.008 | 0.56 | 400 | 0.997 | |
| 28 | Alpha-ketoglutarate | aKG | C00026 | 0.391 | 0.020 | 0.1 | 25 | 0.979 | |
| 29 | Uridine triphosphate | UTP | C00075 | 0.845 | 0.092 | 1.5625 | 400 | 0.998 | |
| 30 | Adenosine triphosphate | ATP | C00002 | 1.557 | 0.188 | 1.5625 | 400 | 0.991 | |
| 31 | Fumarate | FUM | C00122 | 0.576 | 0.100 | 1.5625 | 100 | 0.999 | |
| 32 | Pyruvate | PYR | C00022 | 5.813 | 0.804 | 0.39 | 400 | 0.993 | |
| 33 | Malate | MAL | C00149 | 2.548 | 0.269 | 0.1 | 400 | 0.991 | |
| 34 | D-glyceraldehyde 3-phosphate | GAP | C00118 | 2.194 | 0.367 | 0.1 | 100 | 0.974 | |
| 35 | Acetyl-coenzyme A | ACA | C00024 | 0.196 | 0.044 | 0.1 | 100 | 0.991 | |
| 36 | Nicotinamide adenine dinucleotide phosphate reduced | NADPH | C00005 | 0.006 | 0.010 | 0.14 | 100 | 0.990 | |
| 37 | Phosphoenolpyruvate | PEP | C00074 | 3.442 | 0.345 | 0.1 | 100 | 0.962 | |
| 38 | Succinate | SUCC | C00042 | 5.683 | 0.573 | 0.1 | 320 | 0.999 | |
| 39 | Isocitrate | ICIT | C00311 | 0.003 | 0.006 | 0.39 | 100 | 0.998 | |
| 40 | Citrate | CIT | C00158 | 0.002 | 0.001 | 0.1 | 100 | 0.981 | |
Table 2: Metabolite quantification in a representative CFPS sample. The concentration of each metabolite and the standard deviation. Limit of detection, range of linearity and correlation coefficient identified from standard curves.
Discussion
Cell-free systems have no cell wall, thus there is direct access to metabolites and the biosynthetic machinery without the need for complex sample preparation. However, very little work has been done to develop thorough and robust protocols to quantitatively interrogate cell-free reaction systems. In this study, we developed a fast, robust method to quantify metabolites in cell-free reaction mixtures and potentially in whole-cell extracts. Individual quantification of metabolites in complex mixtures, such as those found in cell-free reactions, or whole-cell extracts, is challenging for several reasons. Central amongst these reasons is chemical diversity. The array of functional groups simultaneously present in these mixtures (e.g., carboxylic acids, amines, phosphates, hydroxyls, etc.) greatly increases the analytical complexity. To circumvent this, we used an aniline derivatization method in combination with 13C internal standards to introduce hydrophobic components to the metabolite mixtures. Using this method, we robustly detected and quantified 40 metabolites in a cell-free reaction in a single LC/MS run. The protocol tagged 35 of the 40 compounds in this study, while the remaining 5 compounds were quantified with a standard curve method. Earlier work suggested the reaction conditions formed an intramolecular salt between the amine and phosphate group that inhibited derivatization12. Reaction conditions were not identified for simultaneous derivatization of all 40 compounds; however, the current alternative is quantification with a standard curve method. While we demonstrated this technique in a cell-free reaction mixture, it could also likely be applied to whole-cell extracts, thus, potentially allowing the absolute quantification of intracellular metabolites concentrations. The latter application has relevance to a variety of important questions in biotechnology and human health.
The method presented here was based on a previous technique (GSIST) that was applied to whole-cell extracts of the yeast S. cerevisiae12,13. In this study, we expanded the number of compounds which could be detected and quantified, including all 12 nucleotides (xMP, xDP, xTP, where x is A, C, G and U). Addition of these compounds could have important biological implications. For example, these nucleotides are heavily involved in transcription and translation processes, which is one of the central processes of interest in CFPS applications, and more generally the compounds are important in a variety of physiological functions. In addition, we were able to detect acetic acid which is an important metabolite when examining overflow metabolism. However, we did not include it in the study because there was a significant reduction of signal in multiple compounds, especially nicotinamide adenine dinucleotide reduced (NADH) and nicotinamide adenine dinucleotide phosphate reduced (NADPH) when acetic acid was added to the standard mixture. Acetic acid had a high limit of detection of 612 µM, thus at these high levels it had a negative effect on the other metabolites’ signals. Despite this, acetic acid can still be detected and quantified in samples by creating a standard curve with just acetic acid in the vial. Acetic acid had a m/z value of 134.0, retention time of 5.78 min, and a linear range from 612 µM to 5000 µM (R2 = 0.986) when tagged with 12C-aniline. The remaining metabolites did not alter each other’s ion signal and represent a comprehensive mixture to characterize CFPS metabolism. The protocol presented here is limited to metabolites involved in central carbon and energy metabolism. Thus, the current method is unable to measure metabolite abundance from other pathways that may be of importance, such as fatty acid and amino acid metabolism.
Taken together, we developed a fast, robust protocol for the characterization and absolute quantification of 40 compounds involved in glycolysis, the pentose phosphate pathway, the tricarboxylic acid cycle, energy metabolism, and cofactor regeneration in CFPS reactions. The method relied on internal standards tagged with 13C-aniline, while the sample was tagged with 12C-aniline. The internal standards and sample compounds co-eluted and eliminated ion-suppression effects which enabled accurate quantification of individual metabolites in complex metabolite mixtures. We identified a total of 40 compounds (41, if including acetic acid) that can be detected and quantified in a cell-free reaction mixture; however, the list of metabolites could be further expanded and adjusted towards the particular biochemical process of interest. Thus, the method provides a robust and accurate approach to characterize cell-free metabolism, which is potentially critical to improving the yield, productivity and energy efficiency of cell-free processes.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The work described was supported by the Center on the Physics of Cancer Metabolism through Award Number 1U54CA210184-01 from the National Cancer Institute ( https://www.cancer.gov/ ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Materials
| 12C Aniline | Sigma-Aldrich | 242284 | Aniline 12C |
| 13C labeled aniline | Sigma-Aldrich | 485797 | Aniline 13C6 |
| 3-Phosphoglyceric acid | Sigma-Aldrich | P8877 | 3PG |
| Acetic Acid | FisherScientific | AC222140010 | ACE |
| Acetonitrile, LCMS | JT BAKER | 9829-03 | ACN |
| Acetyl-coenzyme A | Sigma-Aldrich | A2056 | ACA |
| Acquity UPLC BEH C18 1.7 μM, 2.1 x 150 mm Column | Waters | 186002353 | Column |
| Adenosine diphosphate | Sigma-Aldrich | A2754 | ADP |
| Adenosine monophosphate | Sigma-Aldrich | A1752 | AMP |
| Adenosine triphosphate | Sigma-Aldrich | A2383 | ATP |
| Alpha-ketoglutarate | Sigma-Aldrich | K1128 | aKG |
| Citrate | Sigma-Aldrich | 251275 | CIT |
| Cytidine diphosphate | Sigma-Aldrich | C9755 | CDP |
| Cytidine monophosphate | Sigma-Aldrich | C1006 | CMP |
| Cytidine triphosphate | Sigma-Aldrich | C9274 | CTP |
| D-glyceraldehyde 3-phosphate | Sigma-Aldrich | 39705 | GAP |
| Erythrose 4-phosphate | Sigma-Aldrich | E0377 | E4P |
| Ethanol | Sigma-Aldrich | EX0276 | EtOH |
| Fisher Scientific accuSpin Micro 17 Centrifuge | FisherScientific | Centrifuge | |
| Flavin adenine dinucleotide | Sigma-Aldrich | F6625 | FAD |
| Fructose 1,6-bisphosphate | Sigma-Aldrich | F6803 | F16P |
| Fructose 6-phosphate | Sigma-Aldrich | F3627 | F6P |
| Fumarate | Sigma-Aldrich | F8509 | FUM |
| Gluconate 6-phosphate | Sigma-Aldrich | P7877 | 6PG |
| Glucose | Sigma-Aldrich | G8270 | GLC |
| Glucose 6-phosphate | Sigma-Aldrich | G7879 | G6P |
| Glycerol 3-phosphate | Sigma-Aldrich | G7886 | Gly3P |
| Guanosine diphosphate | Sigma-Aldrich | G7127 | GDP |
| Guanosine monophosphate | Sigma-Aldrich | G8377 | GMP |
| Guanosine triphosphate | Sigma-Aldrich | G8877 | GTP |
| Hydrochloric acid | Sigma-Aldrich | 258148 | HCl |
| Isocitrate | Sigma-Aldrich | I1252 | ICIT |
| Lactate | Sigma-Aldrich | L1750 | LAC |
| Malate | Sigma-Aldrich | 02288 | MAL |
| myTXTL – Sigma 70 Master Mix Kit | ArborBiosciences | 507024 | Cell-free protein synthesis |
| N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride | Sigma-Aldrich | 03449 | EDC |
| Nicotinamide adenine dinucleotide | Sigma-Aldrich | 43410 | NAD |
| Nicotinamide adenine dinucleotide phosphate | Sigma-Aldrich | N5755 | NADP |
| Nicotinamide adenine dinucleotide phosphate reduced | Sigma-Aldrich | 481973 | NADPH |
| Nicotinamide adenine dinucleotide reduced | Sigma-Aldrich | N8129 | NADH |
| Oxalacetate | Sigma-Aldrich | O4126 | OAA |
| Phosphoenolpyruvate | Sigma-Aldrich | P0564 | PEP |
| Pyruvate | Sigma-Aldrich | P5280 | PYR |
| Ribose 5-phosphate | Sigma-Aldrich | R7750 | R5P |
| Ribulose 5-phosphate | CarboSynth | MR45852 | RL5P |
| Sedoheptulose 7-phosphate | CarboSynth | MS07457 | S7P |
| Succinate | Sigma-Aldrich | S3674 | SUCC |
| Tributylamine | Sigma-Aldrich | 90780 | TBA |
| Triethylamine | FisherScientific | O4884 | TEA |
| ultrapure water | FisherScientific | 10977-015 | water |
| Uridine diphosphate | Sigma-Aldrich | U4125 | UDP |
| Uridine monophosphate | Sigma-Aldrich | U6375 | UMP |
| Uridine triphosphate | Sigma-Aldrich | U6625 | UTP |
| VWR Heavy Duty Vortex | VWR | Vortex | |
| Water, LCMS | JT BAKER | 9831-03 | WATER |
| Waters Acquity H UPLC Class Quaternary Solvent Manager | Waters | LCMS | |
| Waters Acquity H UPLC Class Sample Manager FTN | Waters | LCMS | |
| Waters Acquity Qda detector | Waters | LCMS | |
| Waters Empower 3 | Waters | Software | |
| Waters LCMS Total Recovery Vial | Waters | 186000384c | LCMS Vial |
Referencias
- Hodgman, C. E., Jewett, M. C. Cell-free synthetic biology: thinking outside the cell. Metabolic Engineering. 14, 261-269 (2012).
- Vilkhovoy, M., et al. Sequence specific modeling of E. coli cell-free protein synthesis. ACS Synthetic Biology. 7 (8), 1844-1857 (2018).
- Vilkhovoy, M., Minot, M., Varner, J. D. Effective dynamic models of metabolic networks. IEEE Life Sciences Letters. 2 (4), 51-54 (2016).
- Horvath, N., et al. Toward a genome scale sequence specific dynamic model of cell-free protein synthesis in Escherichia coli. bioRxiv. , 215012 (2017).
- Dettmer, K., Aronov, P. A., Hammock, B. D. Mass spectrometry-based metabolomics. Mass spectrometry reviews. 26 (1), 51-78 (2007).
- Hajjaj, H., Blanc, P. J., Goma, G., François, J. Sampling techniques and comparative extraction procedures for quantitative determination of intra- and extracellular metabolites in filamentous fungi. FEMS Microbiology Letters. 164 (1), 195-200 (1998).
- Ruijter, G. J. G., Visser, J. Determination of intermediary metabolites in Aspergillus niger. Journal of Microbiological Methods. 25 (3), 295-302 (1996).
- Mailinger, W., Baltes, M., Theobald, U., Reuss, M., Rizzi, M. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae: I. Experimental observations. Biotechnology and Bioengineering. 55 (2), 305-316 (1997).
- Dunn, W. B., et al. Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics. 9 (1), 44-66 (2013).
- Huang, T., Toro, M., Lee, R., Hui, D. S., Edwards, J. L. Multi-functional derivatization of amine, hydroxyl, and carboxylate groups for metabolomic investigations of human tissue by electrospray ionization mass spectrometry. Analyst. 143 (14), 3408-3414 (2018).
- Huang, T., Armbruster, M. R., Coulton, J. B., Edwards, J. L. Chemical Tagging in Mass Spectrometry for Systems Biology. Analytical Chemistry. 91 (1), 109-125 (2019).
- Yang, W. C., Sedlak, M., Regnier, F. E., Mosier, N., Ho, N., Adamec, J. Simultaneous quantification of metabolites involved in central carbon and energy metabolism using reversed-phase liquid chromatography-mass spectrometry and in vitro 13C labeling. Analytical Chemistry. 80 (24), 9508-9516 (2008).
- Jannasch, A., Sedlak, M., Adamec, J., Metz, T. O. Quantification of Pentose Phosphate Pathway (PPP) Metabolites by Liquid Chromatography-Mass Spectrometry (LC-MS). Metabolic Profiling. Methods in Molecular Biology 708 (Methods and Protocols). , 159-171 (2011).
- Luo, B., Groenke, K., Takors, R., Wandrey, C., Oldiges, M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway, and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. Journal of Chromatography A. 1147 (2), 153-164 (2007).

