Formation of Human Thymus Organoids in Three-Dimensional Fibrin Hydrogels
Summary
Here, we describe a protocol for the formation of human iPSC-derived thymus organoids grown in 3D fibrin hydrogels aiming to support thymic epithelial cell (TEC) maturation and prolonged maintenance, as well as thymopoiesis in vitro.
Abstract
Generation of a functional and self-tolerant T cell repertoire is a complex process dependent on the thymic microenvironment and, primarily, on the properties of its extracellular matrix (ECM). Thymic epithelial cells (TECs) are crucial in thymopoiesis, nurturing and selecting developing T cells by filtering self-reactive clones. TECs have been empirically demonstrated to be particularly sensitive to physical and chemical clues supplied by the ECM and classical monolayer cell culture leads to a quick loss of functionality until their death. Because of this delicate maintenance combined with relative rarity, and despite the high stakes in modeling thymus biology in vitro, models able to faithfully mimic the TEC niche at scale and over time are still lacking. Here, we describe the formation of a multicellular human thymic organoid model, in which the TEC compartment is derived from human induced pluripotent stem cells (iPSC) and reaggregated with primary early thymocyte progenitors in a three-dimensional (3D) fibrin-based hydrogel. This model answers current needs for a scalable culture system that reproduces the thymic microenvironment ex vivo and demonstrates functionality, i.e., the ability to produce T cells and to support thymus organoid growth over several weeks. Thus, we propose a practical in vitro model of thymus functionality through iPSC-derived organoids that would benefit research on TEC biology and T cell generation ex vivo.
Introduction
The thymus is a primary lymphoid organ that plays an essential role in the generation of a competent and tolerant immune system1,2,3. Early thymic progenitors (ETPs) migrate from the bone marrow to the thymus, where they expand and differentiate into functional T cells1,2,4,5. Those processes are mediated by a specialized population, the thymic epithelial cells (TECs)2,6,7. TECs derive from thymic epithelial progenitors (TEPs)8,9 and comprise cortical TECs (cTECs) and medullary TECs (mTECs) that play specific roles in creating the specialized 3D microenvironment needed for T cell migration, expansion, and maturation. TECs mediate T cell development mainly by providing growth and differentiation factors1,10,11 and by negatively selecting non-functional and non-tolerant thymocytes through the presentation of self-antigens5,7,12. The complex interactions between developing T cells and TECs also play a central role in the maturation and 3D organization of the TEC populations in a process known as thymic crosstalk1,11. Interactions between the cell populations of the thymus rely deeply on the specific microenvironment shaped by the extracellular matrix (ECM). The thymic ECM is in a state of dynamic reciprocity with thymic cell populations, impacting gene regulation and being constantly reshaped in return by the secretion of enzymes or matrix proteins13. The ECM influences cells through modification of the bioavailability of growth factors and cytokines, direct signalization through membrane-bound receptors such as integrins, and by shaping cytoskeletons through physical forces14. Thymic ECM components, such as collagens and laminin, have been shown to have a high affinity for growth factors TGFb and FGFs, which are crucial for TEC maintenance and to fix them by forming complexes. Thymic ECM plasticity, elastic modulus, and density also play a crucial role in instructing TEC fate and shaping the compartmentalization of the thymus, which is essential to its functionality. These clues highlight the importance of taking into account the ECM and its 3D structure to mimic the thymus ex vivo. This point is supported by the fact that primary TECs quickly dedifferentiate, lose their functionality, and eventually die when cultivated in classical cell culture setups15,16,17.
Culture models have been developed to expand functional TEC populations from human thymic explants in order to conserve the structure of the ECM and the crucial clues it provides to TECs18,19,20. This culture system was able to successfully expand and maintain a population of functional TECs in vitro but could not be sustained past 7 to 8 days of culture18. Thus, the development of an accessible, practical 3D culture system capable of reproducing the thymic microenvironment and its functionality in vitro and in the long term is a crucial stake in the field. Recently, the development of hydrogel-based 3D culture systems has led to the emergence of several artificial thymic organoid systems, constituting major progress for in vitro thymic modelization15,16,21,22. We developed a human thymic organoid (hTO) co-culture system through the reaggregation of human primary ETPs with human TEPs derived from induced pluripotent stem cells (iPSC) into spheroids and their seeding upon a fibrin hydrogel.
The choice of material and hydrogel setup in this study aimed to reproduce the native structure of the thymic ECM while maintaining practicality and the ability to scale up the process to obtain an affordable and abundant material source for experiments15. This hTO system shows multilineage differentiation potential and can support a productive thymopoiesis from ETPs23. This organoid system constitutes a reliable tool for the study of intrathymic cellular interactions and the modeling of normal and pathological human lymphopoiesis. The use of iPS cells also introduces gene-editing capabilities into the model. Effective differentiation of iPSC into functional thymic tissue has been a longstanding goal of the field for the last 15 years, and significant progress has been made with deciphering TEC lineage fate signalling21,24,25,26,27.To answer the need for such an in vitro 3D thymic model, this technical note describes the methods and technical details for the step-by-step generation of iPSC-derived human thymus organoids, focusing on hydrogel scaffold formation, cell micromass reaggregation and seeding, and organoid culture and harvest.
Protocol
The hiPSC line hiN.Fm.m.Lon71.019 was generated from male adult fibroblasts and reprogrammed via mRNA transfection. The hiPSC line hiN.Fm.f.Lon80.002 was generated from female adult fibroblasts and reprogrammed via mRNA transfection. The hiPSC line hiN.Fs.f.MIPS203.003 was generated from female adult fibroblasts and reprogrammed via recombinant Sendai viral vector infection. All cell lines were provided by the Nantes iPSC platform. Patients gave informed consent for their cells to be used for research purposes (anonymized collection, Lonza, cat # CC-2511). Primary ETPs are isolated by dissociation of postnatal human thymic samples obtained as anonymized discarded waste from patients undergoing pediatric cardiac surgery at Nantes' Hospital (CHU Nantes) the same day, in compliance with the French CODECOH regulation under declaration DC-2017-2987.
1. Directed differentiation of iPSCs towards a TEP identity
NOTE: Since the first works published by Lai and Jin demonstrating the differentiation of murine embryonic stem cells (EScs) towards a thymic epithelial identity28, several studies have developed and optimized protocols describing the directed differentiation of human iPS cells to a TEP identity21,24,25,26,27,29. These studies lead to the differentiation of TEPs expressing thymic epithelial identity markers such as FOXN1 and PAX924,25,28,30, as well as functionality markers such as DLL4 and AIRE26, but lacking TEC maturation markers24,25. Two approaches have been shown to support the maturation of the differentiated TEPs to a mature TEC identity: transplantation into an in vivo model such as mice29, and reaggregation into 3D thymic organoid systems cultivated in an air-liquid interface set-up21. Both systems have demonstrated the crucial role played by the 3D structure in maintaining and supporting the maturation of functional TEC populations capable of supporting in vivo or in vitro T lymphopoiesis15,24,25,31.
- For the thymic organoid system used in this study, conduct the differentiation of iPS cells towards a TEP identity following a protocol developed and detailed in Provin et al.23.
2. Isolation of primary ETPs from a pediatric thymic sample
NOTE: ETPs are bone marrow-originating progenitors giving rise to the T cell lineage and to dendritic cells within the thymus and displaying the following phenotype: CD3- CD4- CD8- CD14- CD19- CD56- CD45+ CD34+ CD7+32,33.
- Preparation of the depletion beads
- The day before, transfer magnetic cell isolation beads (Table of Materials) to a 15 mL tube and wash with 4 mL of Isolation Buffer (PBS + 0.1% BSA + 2 mM EDTA).
- Place the tube in the magnetic stand, remove the supernatant, and add 2 mL of isolation buffer.
- Add mouse anti-human CD3, CD4, and CD8 antibodies to the beads and incubate for 45 min at 4 °C under agitation. Place the tube in the magnetic stand, wash it several times in isolation buffer, and resuspend it in 20 mL of isolation buffer.
- Thymus sample dissociation
- Transfer fresh thymus samples into a petri dish filled with RPMI1640 (Table of Materials). Cut it with sterile dissection scissors and pliers into pieces about 1 mm3 in size.
- With a 25 mL pipette, flush the medium and fragments several times (the medium should turn cloudy), then allow the fragments to sediment and collect half of the medium into a 50 mL tube. Add more medium and repeat until the medium stays clear.
- Collect the medium into as many 50 mL tubes as necessary, and spin the tubes at 200 x g for 5 min.
- Remove the supernatant and resuspend the pellets in 10 mL of red blood cell lysis solution (Table of Materials). Incubate at room temperature (RT) for 5 min and add 20 mL of washing buffer (PBS + 0.5% BSA + 4 mM EDTA + 1% penicillin/streptomycin).
- Spin at 200 x g for 5 min and remove the supernatant. Resuspend the pellets in 10 mL of washing buffer, strain through a 70 µm mesh filter, and count the cells.
- ETP enrichment
- After counting the cells, adjust the volume to 10 mL per tube with the washing buffer. Add the required quantity of cell isolation beads (per 200 million cells, use 500 µL of beads in 20 mL of isolation buffer) and incubate at 4 °C under agitation for 30 min.
- Place the tube on the magnetic stand for 2 min, and carefully collect the supernatant in a clean tube. Remove the tube and clean the beads with 20 mL of isolation buffer. Vortex the tube, place it back on the magnetic stand and collect the supernatant. Repeat this step twice.
- Spin the supernatants at 200 x g for 5 min. Resuspend the pellets in 2 mL of isolation buffer and count the cells.
- ETP isolation
- Adjust the concentration to 200 million cells per mL and collect a small volume as an unstained control.
- Label the cells with mouse anti-human Lineage (Lin) (CD3, CD4, CD8, CD14, CD19, CD56), CD7, and CD34 antibodies (use the same fluorochrome for all Lin markers). Incubate at 4 °C for 45 min.
- Wash the cells in an equal volume of washing buffer. Spin the cells at 200 x g for 5 min, resuspend the pellet in 1 mL, and count the cells.
- Adjust the volume to a concentration of 50 million cells per mL and strain the cells through a 70 µm mesh filter.
- Add the viability marker of choice and sort living Lin- CD34+ CD7+ cells by flow cytometry using a 70 µm nozzle.
3. 3D thymic organoid culture
- TEP preparation
- Immediately after ETP isolation, control the quality of the TEP culture at days 13-15. Ensure that the cells reach confluence and form a dense monolayer with bulges (Figure 1).
- To harvest the differentiated TEPs, wash the cells with DPBS-/-, remove it, add 1 mL of TrypLE (Table of Materials) per well, and incubate at 37 °C for 5-7 min.
- Add 1 mL of XVIVO10 (Table of Materials) per well, flush multiple times to detach the cells, transfer to a 15 mL tube, and spin at 200 x g for 5 min.
- Remove the supernatant, resuspend the pellet in 1 mL of XVIVO10, and count the cells.
NOTE: To assess the differentiation's efficacy ahead of time, use a culture well separately and verify the expression of FOXN1 and PAX9 by RT-qPCR and the differentiation yield by flow cytometry (calculated as the fraction of EPCAM+ CD205+ cells, which should be above 50%) (Figure 1). At this stage of differentiation, nearly all EPCAM+ cells are also positive for CD205, attesting to their precursor identity11.
- ETP preparation
- Immediately after ETP isolation, spin the collection tube at 200 x g for 5 min. Resuspend the pellet in 1 mL of XVIVO10 and count the cells.
- Thymic organoids aggregation
- Pipette the suitable volumes and pool both cell suspensions at a concentration of 2,00,000 TEP and 40,000 ETP per mL, gently pipette up and down once to homogenize.
- Add the suitable supplements according to Table 1, and plate 100 µL of the mixed cell suspension per well in low-binding U bottom 96-well plates. Use a multichannel pipette for increased output; however, a classical single-tip pipette limits losing volumes of precious cells. Incubate the plates at 37 °C and 5% CO2 overnight.
- Preparation of the hydrogels
NOTE: The experimental setup used for hydrogel formation, organoid seeding, and culture medium distribution is shown in Figure 2.- The next day (day 1 of the organoid culture phase), thaw aliquots of Thrombin (10 U/mL), Aprotinin (26,000 U/mL), and Fibrinogen (8 mg/mL) (Table of Materials). Thaw the thrombin and aprotinin on ice and the fibrinogen in a 37 °C water bath (do not put it on ice, as it will precipitate). Do not vortex, but place the aliquots under the hood and homogenize them by gentle pipetting.
- Prepare as many hanging inserts as needed for the number of produced organoids following the ratios presented in Table 2. Place the inserts into the culture wells using sterile pliers, and leave at least one column or row empty in the culture plate.
- Prepare as many 1.5 mL tubes as gels to be cast. In each tube, pipette first the required volumes of Fibrinogen and Aprotinin as detailed in Table 2.
- In a second time, add the required Thrombin volume into a single tube, quickly flush 2 times without creating bubbles to homogenize the reagents, then draw the entire content of the tube and quickly flush the mix into the hanging insert. Position the pipette vertically above the center of the insert, and gently flush the reagent mix without generating bubbles.
NOTE: At this step, speed of execution is crucial as reagents will polymerize in a few seconds, and it is important to mix them correctly to avoid the formation of lumps or uneven density in the gel. Proceed one well at a time using a different 1.5 mL tube for each well (if a tube is reused for several wells, remaining solid gel lumps may block the pipette's tip). If the polymerization occurs too rapidly due to the high activity of the thrombin, use a 1:2 dilution. Just after casting, the gels must be transparent to slightly translucent and still flow after a few seconds when the plate is tilted vertically. - Incubate for at least 1 h at 37 °C until the transparent solution solidifies and turns opaque white, and the gels stay firmly in place when the plate is tilted vertically (Figure 2A).
- Organoid seeding
- Verify the quality of the aggregation step. Ensure that the micromasses form spheric cell masses, with a compact core surrounded by a lower density halo of ETPs (Figure 3).
- Cut the tip of a P200 cone and wash it with an anti-adherence solution (Table of Materials). To harvest the cell masses, tilt the plate to a nearly vertical position: the micromasses will sink to the inferior wall of the wells and can be easily recovered by progressively pushing the pipette tip to the bottom of the well while aspirating.
- Seed the mass at the top of the hydrogels by delicately depositing them without touching the gel with the pipette tip, following the ratios presented in Table 1 (Figure 2B). Even if organoids seem free-floating at this stage, when swollen with culture medium, the gel will soften, and the organoids will nestle in the upper layer. Check under the microscope that no organoid is left in the P96 wells.
- Prepare the required volume of culture medium according to Table 1 and Table 2, and in each well, slowly add a quarter of the volume at the top of the hydrogels without touching them by pipetting along the insert walls and the remaining three-quarters at the bottom of the well by positioning the pipette in between the arms of the hanging insert (Figure 2C).
- Place 1 mL of PBS in the empty culture wells to maintain humidity in the plate. Incubate at 37 °C and 5% CO2.
- Thymic organoid culture
- On day 2, check if the organoids are well seeded: the hydrogels must have stayed in place, and the organoids must not have sedimented at the bottom of the insert.
- Prepare the required amount of culture medium following Table 1 and Table 2. Remove the medium by directing the tip of the aspiration cone between the arms of the hanging insert, making sure not to touch the gel. Add the new medium by positioning the pipette in the same manner.
- Change the medium every 2 days, switching to the second phase medium (Table 1) after 2 to 4 days (on day 18 following the start of the TEP differentiation).
NOTE: The organoid batch can be maintained in culture in this manner for up to 6 weeks.
- Organoid harvesting
- Prepare a 15 mL tube with 1 mL of TrypLE per well to harvest.
- Cut the tip of a P1000 cone and coat it in an anti-adherence solution. Gently pipette the gel by placing the pipette tip vertically at the center of the inserts (be careful not to perforate the membrane) and transfer to the TrypLE tube. Wash the insert's membrane with TrypLE and transfer to the tube as well.
- Incubate at 37 °C for 15 min, vortexing gently at 5 min intervals. Ensure that the gels and organoids dissociate.
- After 15 min, strain on a 70 µm mesh filter and spin at 200 x g for 5 min. Resuspend the pellet in the washing buffer and proceed to the analytical method of choice.
Representative Results
The workflow of the protocol is summarized in Figure 4. For this 3D organoid culture model, we adopted a thrombin and fibrinogen hydrogel that had previously been used by our team to maintain primary mouse mTECs for a couple of days, thanks to the physical and mechanical cues it provided34. After polymerization, the gel should display a loose, sponge-like mesh structure (Figure 5).
After the initial seeding and attachment phase, the organoids progressively grew and developed both at the surface and within the uppermost layers of the gel. Depending on the gel properties, the seeding conditions, and the number of organoids seeded on the gel, the organoids formed spheric to oblong structures (Figure 6) and occasionally merged to form larger structures. Two particular sub-levels of the organization were observed within the organoids after the first week of culture: first, we observed long, cell-surface projection-like structures formed by large cells irradiating from the organoids and colonizing the hydrogel in all directions (Figure 6 and Figure 7). Second, we observed cluster-like structures formed by smaller cells concentrating around those cell projections. Although we were not able to isolate both cell types to confirm the study hypothesis, this phenomenon is reminiscent of 3D arrangements found within the thymic cortex, formed by the interaction of individual cTECs with a large number of much smaller developing T cells, known as thymic nurse cell complexes11 (Figure 8).
At several time points during the organoid culture phase, we evaluated the cellular composition of the thymic organoids by flow cytometry and identified several key compartments: TEC (characterized as EPCAM+ CD45-), thymocytes (EPCAM- CD45+ CD3+) (Figure 9), as well as an EPCAM- CD45+ CD3- compartment comprising thymic hematopoietic non-thymocyte subsets. Further details can be found in Provin et al.23.
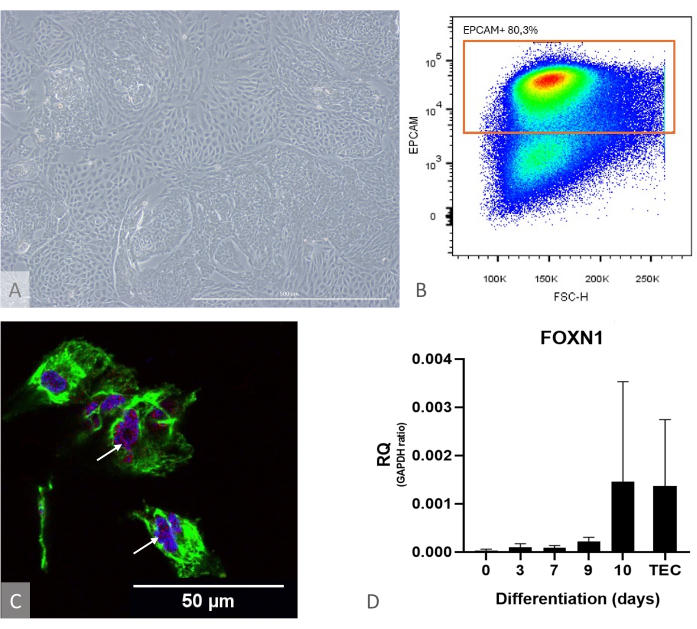
Figure 1: iPSC to TEP differentiation characterization. (A) Example of iPSC to TEP differentiation at D13, inverted phase contrast microscope, 400x. Scale bar: 500 µm. (B) Dot plot example, the proportion of EPCAM+ cells among DAPI- cells on day 14 of differentiation, image from FlowJo 10.0.7. (C) Immunostaining against DAPI (blue), PAX9 (red), and KRT8 (green), immunofluorescence, and confocal imaging on day 16 of iPSC to TEP differentiation. White arrows point to examples of anti-PAX9 staining. Scale bar: 50 µm (D) Expression level of FOXN1 (RQ to GAPDH) during iPS to TEP differentiation. TEC: Positive control reference, primary human TECs isolated from pediatric thymus samples. Graph from Prism (GraphPad version 8.0.1). Please click here to view a larger version of this figure.
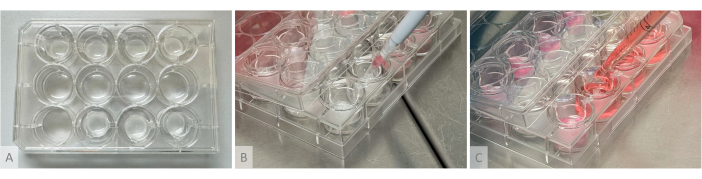
Figure 2: Experimental setup for hydrogel formation, organoid seeding, and culture medium distribution. (A) Culture plate with hydrogels casted into hanging inserts placed in the top and bottom rows. (B) Organoid seeding: the cut pipette cone containing 1 organoid is placed above the hydrogel without touching it, and the organoid is gently seeded at the surface of the gel. (C) Culture medium is deposited in the culture well by positioning the tip of the pipette between the arms of the hanging insert. Please click here to view a larger version of this figure.
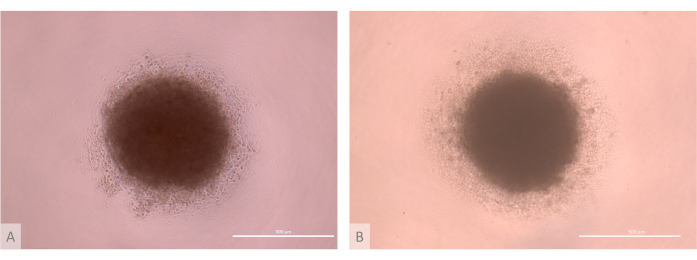
Figure 3: D0 of thymic organoid culture before seeding (days 13-15 of the full protocol). (A) organoid produced with TECs derived from the Lon71.019 iPS line. (B) Organoid produced with TECs derived from the MIPS203.003 iPS line. Inverted phase contrast microscope, 1000x. Scale bars: 500 µm. Please click here to view a larger version of this figure.
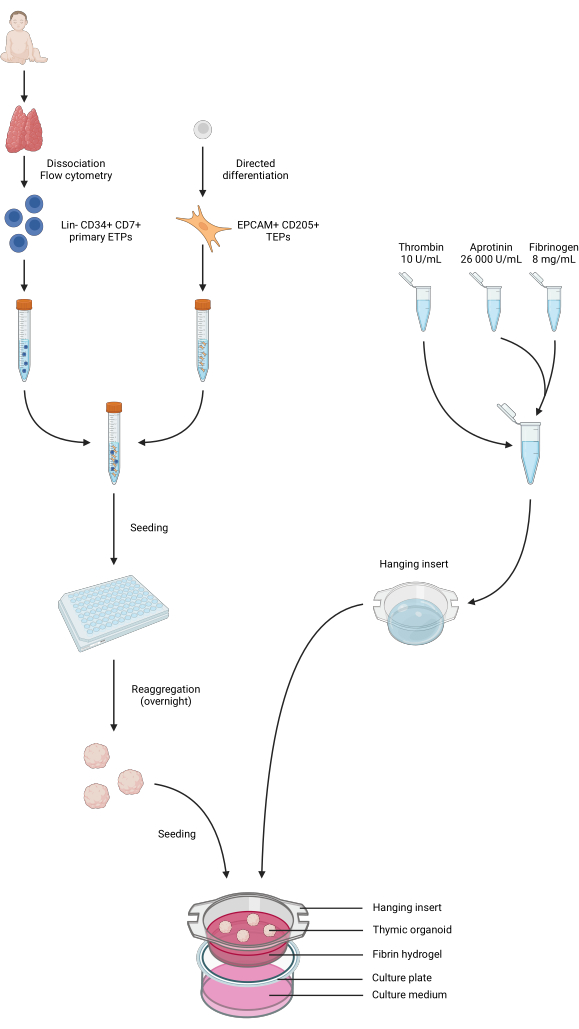
Figure 4: Summarized representation of all steps of the protocol. Pediatric thymus samples were collected and dissociated, and primary Lin- CD34+ CD7+ ETPs were sorted by flow cytometry. Differentiation of iPS cells was conducted towards a TEP identity. ETPs and iPS-derived TEPs were pooled and seeded in low-binding 96-well plates and aggregated into thymic organoids overnight. Fibrin hydrogels were prepared from aprotinin, fibrinogen, and thrombin and casted into hanging inserts. After polymerization, the organoids were seeded atop the hydrogels, and the phase 1 culture medium was added to the wells. The organoids were kept in culture for up to 6 weeks. Created in BioRender, publication license AG26EFCZOM. Please click here to view a larger version of this figure.
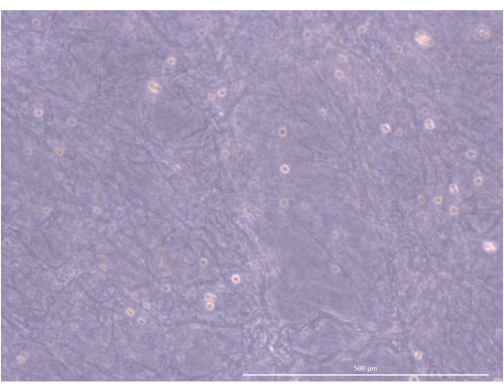
Figure 5: Organization and structure of the hydrogel. Inverted phase contrast microscope, 1000x. Scale bar: 500 µm. Please click here to view a larger version of this figure.
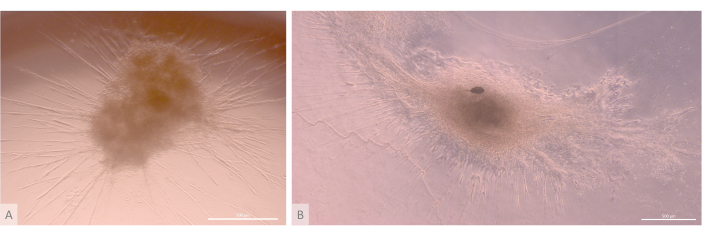
Figure 6: Mature organoids and three-dimensional structure. (A) Thymic organoid on day 24 of 3D culture, MIPS203.003 iPS line. (B) Composite image of a thymic organoid on day 32 of 3D culture, Lon71.019 iPS line. Inverted phase contrast microscope. Scale bars: 500 µm. Please click here to view a larger version of this figure.
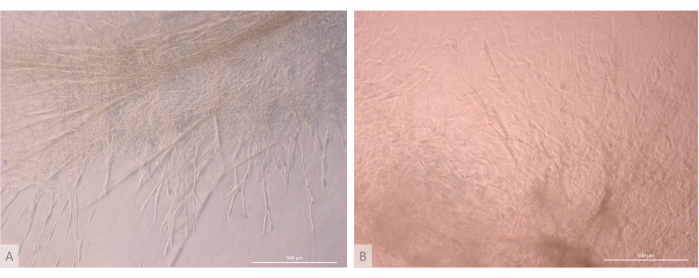
Figure 7: Structure detail of thymic organoids. (A) Thymic organoid on day 32 of 3D culture, L71.019 iPS line. (B) Thymic organoid on day 27 of 3D culture, L80.002 iPS line. Inverted phase contrast microscope, 400x. Scale bars: 500 µm. Please click here to view a larger version of this figure.
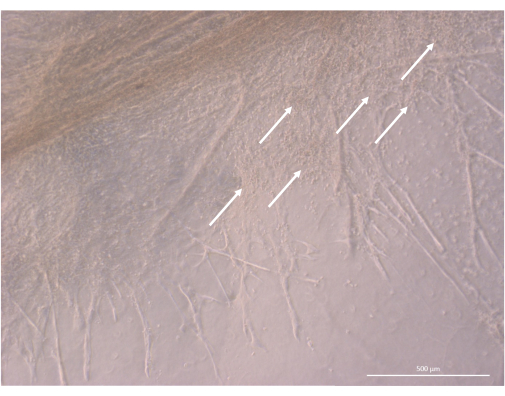
Figure 8: Structure detail of a thymic organoid on day 32 of 3D culture. White arrows point to clusters of small thymocytes proliferating in close proximity to TEC cells. Inverted phase contrast microscope, 400x. Scale bar: 500 µm. Please click here to view a larger version of this figure.
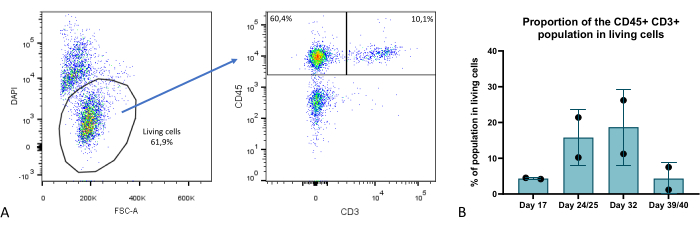
Figure 9: Proportion of the T cell compartment within thymic organoids. (A) Dot plot example, proportion of CD45+ CD3+ cells within the living (DAPI-) cells in thymic organoids on day 35 of 3D culture, image from FlowJo 10.0.7. The CD45+ CD3- fraction comprises hematopoietic non-thymocyte cells. (B) The proportion of CD45+ CD3+ cells within living cells in thymic organoids on days 17, 24/25, 32, and 39/40 of 3D culture, n=2 in technical duplicate or triplicate, graph from Prism (GraphPad version 8.0.1). Please click here to view a larger version of this figure.
| Unit | Phase 1 medium Day 14 up to Day 18 | Phase 2 medium Day 19 onward | |
| Base | XVIVO10 | XVIVO10 | |
| BMP4 | ng/mL | 50 | |
| FGF8 | ng/mL | 10 | |
| FGF10 | ng/mL | 10 | |
| IGF1 | ng/mL | 10 | |
| EGF | ng/mL | 10 | |
| RANK L | ng/mL | 50 | 50 |
| IL7 | ng/mL | 5 | 5 |
| FLT3 L | ng/mL | 5 | 5 |
| SCF | ng/mL | 10 | 10 |
| Glutamax | ng/mL | 1% | 1% |
Table 1: Supplements and their respective concentrations.
| Aprotinin (µL) | Thrombin (µL) | Fibrinogen (µL) | Phase 1 medium | Organoids (unit) | |
| 24 well plate | 5 | 75 | 75 | 1 | 3 to 5 |
| 12 well plate | 9.2 | 138.2 | 138.2 | 1.8 | 5 |
| 6 well plate | 16 | 240.8 | 240.8 | 3.2 | 8 to 9 |
Table 2: Required ratios of components for preparing hydrogels and seeding organoids in 6-, 12- and 24-well plates.
Discussion
Compared with classical monolayer culture in 2D or even more advanced state-of-the-art 3D models such as RTOC (reaggregated thymus organ culture), the model we describe here presents significant improvements. From a technical point of view, this model offers improved scalability and reproducibility as TECs are derived from self-renewing iPS cells. It also allows gene editing at the iPSC stage for easier knock in or knock out studies in TECs. The survivability of the thymic organoids shown in this study is remarkable and provides a significant improvement comparatively to 2D or RTOC cultures, with demonstrated T cell generation during up to 6 weeks (Figure 9). Thus, the reconstitution of the thymic 3D structure and ECM properties leads to sustained thymic functionality in our thymic organoids, i.e., the ability to generate T cells from the most mature thymocyte compartment, recent thymic emigrants at around week 4 of 3D culture, with generation of both CD4+ and CD8+ T cells23.
Because the thymic microenvironment supports an intense expansion and differentiation activity, proper gaseous exchange is a crucial parameter in any in vitro thymus model. Indeed, improved results have been observed in models maintained either in an enriched dioxygen atmosphere or at air-liquid interfaces21,35. Our observations support this point and highlight the importance of a correct organoid seeding at the top of the hydrogel just under the air interface. Defects in polymerization leading to viscous to liquid hydrogels will cause the sinking of organoids at the bottom of the inserts and hinder their growth. Coculture with endothelial cells on-chip is a promising alternative that could break this barrier by adding vascularization. The size of the thymus organoids produced in this study is limited to around 5 mm, allegedly due to a lack of gas and nutrient exchanges in the core areas. Vascularization would thus allow culture scale-up and, combined with process optimization, permit the production of organoids containing millions of TECs and T cells. The density of the hydrogel is also a crucial parameter, and its reproducibility across batches is one of the main limitations of the protocol, given the enzymes' sensibility to freezing and thawing cycles. The hydrogel casting step is a critical step in the protocol; we recommend performing a test by casting one hydrogel 1 h before any planned experiment to check reagent activity. In case of insufficient enzymatic activity leading to impaired polymerization and given the cost of the iPSC-derived TEPs, we advise no other troubleshooting than starting the protocol again with fresh reagents aliquots. TECs are important producers of ECM; however, given the recent advances in the understanding of the role of thymic fibroblasts, it could be interesting to add a population of irradiated fibroblasts into the organoid model. This population could secrete growth factors and ECM that would participate in reproducing the thymic environment with positive effects on TEC and T cell differentiation and maintenance. Another important limitation of this thymus organoid model is the lack of proper cortico-medullary segregation. Because the capsular fibroblasts of the thymus have been shown to shape the formation of the cortex, their addition to the culture model could help address this limitation. Thus, this protocol introduces the basis of complex in vitro models of the thymus. It combines the recent advances made in the fields of iPSC thymic differentiation, 3D hydrogel-based cultures, and in vitro lymphopoiesis. This model can be further refined to address scalability and increase its complexity, for instance, by adding mesenchymal and vascular compartments. It could thus result in valuable research platforms on immunity or applications in personalized T-cell-based cell therapy.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We want to thank the members of the iPSC core facility of Nantes, France, headed by Laurent David. This work was supported by the JP-Rare Disease JTC2019 program TARID project (EJPRD19-208) funded by the ANR (ANR-19-RAR40011-5) to M.G. by the RFI Bioregate grant (ThymIPS) from la Région Pays de la Loire to M.G., by the ANR (ANR-22-CE15-0045) to M.G. and the "SATT Ouest Valorisation" project OrgaTreg to M.G. N.P. was supported by "la fondation d'entreprise ProGreffe". M.d.A. was supported by "la Fondation pour la Recherche Médicale". We thank the iPSC core facility of Nantes, supported by IBiSA and Biogenouest, for the use of their resources and technical support. This work was partially funded by the Labex IGO program supported by the National Research Agency via the investment of the future program ANR-11-LABX-0016-01.
Materials
| Aprotinin | Sigma Aldrich | 616370 | |
| BMP4 | Miltenyi | 130-111-165 | |
| CCR7 (CD197) | BD Biosciences | PE | Clone: 3D12; Dilution: 1: 200 |
| CD14 | BD Biosciences | FITC | Clone: M5E2; Dilution: 1: 200 |
| CD19 | BD Biosciences | PE | Clone: HIB19; Dilution: 1: 200 |
| CD205 | BioLegend | FITC | Clone: MG38; Dilution: 1: 200 |
| CD3 | BD Biosciences | PE | Clone: HIT3a; Dilution: 1: 200 |
| CD34 | BD Biosciences | FITC | Clone: 8G12; Dilution: 1: 100 |
| CD4 | BD Biosciences | PE | Clone: RPA-T4; Dilution: 1: 100 |
| CD4 | BD Biosciences | BV711 | Clone: L200; Dilution: 1: 200 |
| CD45 | BD Biosciences | PerCP | Clone: HI30; Dilution: 1: 200 |
| CD56 | BD Biosciences | PE | Clone: B159; Dilution: 1: 200 |
| CD62L | BD Biosciences | BV605 | Clone: DREG-56; Dilution: 1: 200 |
| CD69 | BD Biosciences | BV510 | Clone: FN50; Dilution: 1: 200 |
| CD7 | BD Biosciences | APC | Clone: M-T701; Dilution: 1: 200 |
| CD8 | BD Biosciences | PeCy7 | Clone: RPA-T8; Dilution: 1: 200 |
| CD8 | BD Biosciences | PE | Clone: HIT8a; Dilution: 1: 200 |
| Dynabeads Pan Mouse IgG | Invitrogen | 11041 | |
| EGF | Miltenyi | 130-097-751 | |
| EPCAM (CD326) | BD Biosciences | PE | Clone: HEA-125; Dilution: 1: 200 |
| EPCAM (CD326) | Miltenyi | BV711 | Clone: EBA-1; Dilution: 1: 200 |
| FGF10 | Miltenyi | 130-127-858 | |
| FGF8 | Biotechne R&D | 423-F8 | |
| Fibrinogen | Sigma Aldrich | 341578 | |
| FLT3 L | Peprotech | AF-300-19 | |
| Glutamax | Gibco | 35050-61 | |
| IGF1 | Miltenyi | 130-093-886 | |
| IL7 | Peprotech | AF-200-07 | |
| RANK L | Biotechne R&D | 6449-TEC | |
| Red blood cell lysis solution | Miltenyi | 130-094-183 | |
| RPMI1640 | Gibco | 11875093 | |
| SCF | Peprotech | AF-300-07 | |
| Thrombin | Sigma Aldrich | 605190 | |
| TrypLE | Gibco | 2605010 | |
| XVIVO10 | Lonza | LONBE04-380Q |
Referenzen
- Starr, T. K., Jameson, S. C., Hogquist, K. A. Positive and negative selection of T cells. Annu Rev Immunol. 21, 139-176 (2003).
- Carpenter, A. C., Bosselut, R. Decision checkpoints in the thymus. Nat Immunol. 11 (8), 666-673 (2010).
- Miller, J. F. A. P. The function of the thymus and its impact on modern medicine. Science. 369 (6503), (2020).
- Haddad, R., et al. Dynamics of thymus-colonizing cells during human development. Immunity. 24 (2), 217-230 (2006).
- Cumano, A., et al. New molecular insights into immune cell development. Annu Rev Immunol. 37, 497-519 (2019).
- Bautista, J. L., et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat Commun. 12 (1), 1096 (2021).
- Kadouri, N., Nevo, S., Goldfarb, Y., Abramson, J. Thymic epithelial cell heterogeneity: TEC by TEC. Nat Rev Immunol. 20 (4), 239-253 (2020).
- Alves, N. L., et al. Serial progression of cortical and medullary thymic epithelial microenvironments. Eur J Immunol. 44 (1), 16-22 (2014).
- Baik, S., Jenkinson, E. J., Lane, P. J. L., Anderson, G., Jenkinson, W. E. Generation of both cortical and Aire+ medullary thymic epithelial compartments from CD205+ progenitors. Eur J Immunol. 43 (3), 589-594 (2013).
- Tavian, M., Peault, B. Embryonic development of the human hematopoietic system. Int J Dev Biol. 49 (2-3), 243-250 (2005).
- Abramson, J., Anderson, G. Thymic epithelial cells. Annu Rev Immunol. 35 (1), 85-118 (2017).
- Anderson, M. S., et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 298 (5597), 1395-1401 (2002).
- Sharma, H., Moroni, L. Recent advancements in regenerative approaches for thymus rejuvenation. Adv Sci. 8 (14), 2100543 (2021).
- Alenghat, F. J., Ingber, D. E. Mechanotransduction: all signals point to cytoskeleton, matrix, and integrins. Sci STKE. 2002 (119), 6 (2002).
- Pinto, S., Schmidt, K., Egle, S., Stark, H. -. J., Boukamp, P., Kyewski, B. An organotypic coculture model supporting proliferation and differentiation of medullary thymic epithelial cells and promiscuous gene expression. J Immunol. 190 (3), 1085-1093 (2013).
- Hun, M., Barsanti, M., Wong, K., Ramshaw, J., Werkmeister, J., Chidgey, A. P. Native thymic extracellular matrix improves in vivo thymic organoid T cell output, and drives in vitro thymic epithelial cell differentiation. Biomaterials. 118, 1-15 (2017).
- Asnaghi, M. A., et al. Thymus extracellular matrix-derived scaffolds support graft-resident thymopoiesis and long-term in vitro culture of adult thymic epithelial cells. Adv Funct Mater. 31 (20), 2010747 (2021).
- Villegas, J. A., et al. Cultured human thymic-derived cells display medullary thymic epithelial cell phenotype and functionality. Front Immunol. 9, 1663 (2018).
- Hauri-Hohl, M., Zuklys, S., Holländer, G. A., Ziegler, S. F. A regulatory role for TGF-β signaling in the establishment and function of the thymic medulla. Nat Immunol. 15 (6), 554-561 (2014).
- Campinoti, S., et al. Reconstitution of a functional human thymus by postnatal stromal progenitor cells and natural whole-organ scaffolds. Nat Commun. 11 (1), 6372 (2020).
- Ramos, S. A., et al. Generation of functional thymic organoids from human pluripotent stem cells. Stem Cell Reports. 18 (4), 829-840 (2023).
- Fan, Y., et al. Bioengineering thymus organoids to restore thymic function and induce donor-specific immune tolerance to allografts. Mol Ther. 23 (7), 1262-1277 (2015).
- Provin, N., et al. Combinatory differentiation of human induced pluripotent stem cells generates thymic epithelium that supports thymic crosstalk and directs dendritic- and CD4/CD8 T-cell full development. bioRxiv. 2023, 572664 (2023).
- Parent, A. V., et al. Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development. Cell Stem Cell. 13 (2), 219-229 (2013).
- Sun, X., et al. Directed differentiation of human embryonic stem cells into thymic epithelial progenitor-like cells reconstitutes the thymic microenvironment in vivo. Cell Stem Cell. 13 (2), 230-236 (2013).
- Inami, Y., et al. Differentiation of induced pluripotent stem cells to thymic epithelial cells by phenotype. Immunol Cell Biol. 89 (2), 314-321 (2011).
- Gras-Pena, R., et al. Human stem cell-derived thymic epithelial cells enhance human T cell development in a xenogeneic thymus. J Allergy Clin Immunol. 149 (5), 1755-1771 (2022).
- Lai, L., Jin, J. Generation of thymic epithelial cell progenitors by mouse embryonic stem cells. Stem Cells. 27 (12), 3012-3020 (2009).
- Ramos, S. A., et al. Generation of functional human thymic cells from induced pluripotent stem cells. J Allergy Clin Immunol. 149 (2), 767-781 (2022).
- Provin, N., Giraud, M. Differentiation of pluripotent stem cells into thymic epithelial cells and generation of thymic organoids: Applications for therapeutic strategies against APECED. Front Immunol. 13, 930963 (2022).
- Montel-Hagen, A., et al. In vitro recapitulation of murine thymopoiesis from single hematopoietic stem cells. Cell Rep. 33 (4), 108320 (2020).
- Park, J. -. E., et al. A cell atlas of human thymic development defines T cell repertoire formation. Science. 367 (6480), 3224 (2020).
- Flippe, L., et al. Rapid and reproducible differentiation of hematopoietic and T cell progenitors from pluripotent stem cells. Front Cell Dev Biol. 8, 577464 (2020).
- Padonou, F., et al. Aire-dependent transcripts escape Raver2-induced splice-event inclusion in the thymic epithelium. EMBO Rep. 23 (3), e53576 (2022).
- Han, J., Zúñiga-Pflücker, J. C. High-oxygen submersion fetal thymus organ cultures enable FOXN1-dependent and -independent support of T lymphopoiesis. Front Immunol. 12, 652665 (2021).

