An Intravital Microscopy-Based Approach to Assess Intestinal Permeability and Epithelial Cell Shedding Performance
Summary
Taking advantage of intravital microscopy, the method presented here enables real-time visualization of intestinal epithelial cell shedding in living animals. Therefore, topically stained intestinal mucosa (acriflavine and rhodamineB-dextran) of anesthetized mice is imaged up to single-cell resolution using confocal microscopy.
Abstract
Intravital microscopy of the gut using confocal imaging allows real time observation of epithelial cell shedding and barrier leakage in living animals. Therefore, the intestinal mucosa of anesthetized mice is topically stained with unspecific staining (acriflavine) and a fluorescent tracer (rhodamine-B dextran), mounted on a saline solution-rinsed plate and directly imaged using a confocal microscope. This technique can complement other non-invasive techniques to identify leakage of intestinal permeability, such as transmucosal passage of orally administered tracers. Besides this, the approach presented here allows the direct observation of cell shedding events at real-time. In combination with appropriate fluorescent reporter mice, this approach is suitable for shedding light into cellular and molecular mechanisms controlling intestinal epithelial cell extrusion, as well as to other biological processes. In the last decades, interesting studies using intravital microscopy have contributed to knowledge on endothelial permeability, immune cell gut homing, immune-epithelial communication and invasion of luminal components, among others. Together, the protocol presented here would not only help increase the understanding of mechanisms controlling epithelial cell extrusion, but could also be the basis for the developmental of other approaches to be used as instruments to visualize other highly dynamic cellular process, even in other tissues. Among technical limitations, optical properties of the specific tissue, as well as the selected imaging technology and microscope configuration, would in turn, determine the imaging working distance, and resolution of acquired images.
Introduction
The intestine is a highly specialized organ with a tightly regulated function enabling conflicting processes, namely nutrition and protection against harmful luminal substances. Lining between the human body and the environment, the intestinal epithelium acts as a physical and immunological barrier and contributes to the maintenance of mucosal homeostasis in the gut1,2. Loss of epithelial integrity and increased tight junction permeability is well known to be associated with Inflammatory Bowel Disease (IBD)3,4,5,6. Epithelial alterations are then considered as causes and secondary amplifiers for chronic intestinal inflammation in IBD. Thus, an improved understanding of early epithelial alterations in the gut of IBD patients would be of immense value for the development of new strategies to restore epithelial integrity for reliable prediction and subsequent prevention of IBD relapses.
Intestinal epithelium follows a complex and tightly regulated turnover process. From the crypt bottom, terminally differentiated intestinal epithelial cells (IECs) derived from pluripotent stem cells migrate upwards to the villus tip, where aged/damaged cells are shed into the lumen7. The equilibrium between division and cell extrusion enables the maintenance of intestinal epithelial cell numbers, avoiding the formation of gaps and leakage, as well as the accumulation of epithelial cells potentially leading to cell masses and tumorigenesis8,9,10. Despite the key role of epithelial cell shedding in the physiological renewal of the gut epithelium, the knowledge about the molecular mechanisms driving the extrusion of cells at the villus tip is limited. Thus, there is a need for basic research providing a precise description of the sequence of molecular events involved in epithelial cell shedding.
Complex interactions between different cell types within the intestinal mucosa are key to understand the molecular mechanisms regulating epithelial turnover and intestinal homeostasis. Thus, in vivo studies offer high advantages over in vitro and ex vivo approaches in this context. Moreover, real-time imaging techniques permit the description of the sequence of events controlling specific phenomena. In this context, the study of highly dynamic processes demands the use of optimized high resolution techniques for the direct observation of the tissue. Intravital imaging techniques appear as unique suitable tools for the study of epithelial cell shedding in the gut.
The term "intravital microscopy" refers to experimental approaches taking advantage of high-resolution imaging techniques (multiphoton or confocal microscopy) to directly visualize cells and tissues in their native environs within a living animal11. It enables real time acquisition of in vivo information up to single-cell resolution, and entails clear advantages over static or low resolution methods. Intravital microscopy provides complementary information and overcome some limitations from classical and/or high-end techniques, such as artifacts due to tissue processing. In contrast, the main limitation of intravital microscopy is that the tissue should be directly exposed to the microscope, which in most cases requires surgery. Although sophisticated approaches preserve the vitality and minimize the impact of the imaged tissue (skinfold chambers and imaging windows)12,13, in most cases a simple skin incision is performed for the externalization of the tissue (skin flaps)14. In the last decade, these approaches have contributed key evidence about highly dynamic processes, which were previously inscrutable. Translationally, real-time imaging provided new biological insights on stem cell and leukocytes homing15, as well as cancer dissemination and metastasis formation13,16. In the clinical context, endomicroscopy is currently exploited as a diagnostic tool of cancer17 and gastrointestinal diseases, such as IBD18,19; while confocal mosaicking microscopy became a rapid pathology tool during surgery20. Together, intravital microscopy has lately emerged as a valuable and versatile tool for biomedical research and future application in the clinic.
Intravital microscopy is here implemented for real-time visualization of intestinal epithelial leakage and observation of epithelial cell shedding events. Leakage of intestinal permeability can be identified by other in vivo noninvasive techniques, such as quantification of orally administration of fluorescent tracers in serum21. However, this technique does not allow the direct observation of shedding performance nor the segregation between para- and trans-cellular permeability. The combination of standard tracer experiments and intravital microscopy represents a suitable approach to: i) identify disturbances in intestinal permeability, and ii) segregate between para- and trans-cellular epithelial permeability. Besides cell shedding, intravital microscopy in combination with in vivo fluorescence labelling enables the study of other cellular and molecular mechanisms (e.g., tight junction redistribution during cell shedding using fluorescent reporter mice22 or interactions between IECs and other cells within the intestinal mucosa23).
The method presented here represents an adaptation of intravital microscopy to enable real-time observation of intestinal mucosa, using confocal laser scanning microscopy (CLSM). Therefore, we use conditional knock-out mice of GGTase (Geranylgeranyltransferase) in intestinal epithelial cells (IECs) in (Pggt1biΔIEC mice), since they suffer from a severe intestinal disease and increased epithelial permeability24. Surgical preparation of the mouse and staining of the intestinal mucosa, as well as appropriate settings used for imaging acquisition and post-acquisition analysis are described. This protocol could enable future studies contributing to the current knowledge about dynamics and kinetics of intestinal epithelial cell shedding. Moreover, the protocol could serve as a basis for various adaptations to study other phenomena occurring at the surface of the intestinal mucosa, and even at other tissues.
Protocol
The following protocol has been approved by the relevant local authorities in Erlangen (Regierung von Unterfranken, Würzburg, Germany). Mice were housed under specific pathogen-free conditions.
NOTE: Inhibition of GGTase-mediated prenylation within IECs causes a severe alteration of intestinal permeability in Pggt1biΔIEC mice24. Therefore, this mouse model was used to demonstrate how the protocol can be useful to study intestinal barrier defects. However, this protocol could be used for the study of any other mouse line.
1. Surgical preparation and mouse intestinal mucosa staining
NOTE: The surgical preparation is based on previously described protocols25. Keep the anesthetized mouse under a red lamp during the surgical preparation, to avoid a drop in body temperature.
- Anesthetize mouse by intraperitoneal injection of ketamine/xylazin (96 mg/kg ketamine; and 12.8 mg/kg xylazin). Verify the anesthesia by checking for the lack of an eyelid reflex.
- Apply eye protection cream using a cotton bud.
- Make an incision (1 cm) on the left ventral area using standard forceps and straight fine scissors.
- Exteriorize a segment of the intestine (5-7 cm, approximately).
- Open the exteriorized intestinal segment longitudinally by electrocauterization at the antimesenteric side.
- Expose the mucosa and rinse shortly with saline solution to remove fecal content.
NOTE: Optionally, apply xylazin directly on the gut to avoid motion artifacts due to peristalsis. - Stain the surface of the intestinal mucosa with acriflavine and rhodamine-B dextran (10 kDa).
- Apply the 1 mg/mL acriflavine solution (100 µL) by pipetting drop by drop on the mucosa and incubate for 3 min. Wash out the remaining solution with PBS.
- Apply the 2 mg/mL rhodamine-dextran solution (100 µL) by pipetting drop by drop on the mucosa and incubate for 3 min. Wash out the remaining solution with PBS.
NOTE: Optionally, remove blood from the preparation using aseptic cotton.
- Place the anesthetized mouse supine on a cover slide mounted in a chamber rinsed with pre-warmed saline solution (37 °C).
NOTE: The opened intestinal segment is then placed luminal surface down, on the cover slide. - Place the preparation (anesthetized mouse in the chamber) on the inverted microscope stage.
- Cover the animal with an isothermal pad (approx. 37 °C).
- Proceed immediately to intravital microscopy.
NOTE: Keep the surgical preparation rinsed with pre-warmed saline solution to avoid dehydration of the tissue and cell death.
2. Intravital microscopy
NOTE: Perform step 2.1 before starting the surgical preparation, to avoid long waiting times between anesthesia, surgery and image acquisition. If necessary, additional doses of anesthetics can be given to surgically prepared animals to keep them under anesthesia for the imaging experiments.
- Set up the CLSM microscope.
- Start the CLSM microscope by turning on the microscope base and the scanner box. Turn on the computer by pressing the Start button.
- Launch the image acquisition software (e.g., LAS X) by double clicking on the icon. Select the appropriate configuration (Configuration: machine; Microscope: DMI6000) and click OK.
- Define the appropriate Resolution. Go to Configuration | Hardware | Resolution | Bit depth. Select 12.
- Go to the Acquisition menu. Select xyzt for the image acquisition mode from the drop-down menu. Select the objective from the drop-off menu (20x or 40x).
- Design the sequential acquisition setting. Click on Seq. Add a second sequence, by clicking on the add button (+). Select Between frames.
- Configure Sequence 1 (detection of acriflavine; 416 nm excitation; 514 nm emission). Turn on the visible laser box. Activate the PMT1 (ON). Define the emission wavelength window (490-550 nm).
- Configure Sequence 2 (detection of rhodamineB-dextran; 570 nm excitation; 590 nm, emission). Turn on the visible laser box. Activate the PMT2 (ON). Define the emission wavelength window (550-760).
- Activate the corresponding lasers (488 and 552). Go to Configuration | Lasers. Activate 488 and 552 nm lasers (turn ON).
- Adjust the setting to the specific features of the current experiment.
- Turn ON the light source (press power). Place the preparation (anesthetized mouse in the chamber) on the microscope stage and change the xy position until the illumination axis is focused on the tissue preparation.
- Select the field of interest using the standard light source and the eyepieces.
- Select the filter cube (I3). Open the shutter. Focus on the surface of the intestinal mucosa by using the macro- and micro-wheel. Search for an area where several villi can be visualized within the field of view by changing the XY position.
- Verify that the rhodamine-dextran staining is also visible in that area. Change the filter cube (N2.1). Check the image through the eyepieces.
- Start the CLSM image acquisition from the software. Optimize settings for the two sequences.
- Select Sequence 1. Adjust laser power, gain and offset for Sequence 1.
- Select Sequence 2. Adjust laser power, gain and offset for Sequence 2.
- Define the z stack range.
- Open the Z-stack drop-off menu. Focus on the surface of the mucosa using the z-axis control. Press Begin.
- Focus on the bottom limit where the signal is still detectable. Press End.
- Define the numbers of z stacks (10). Avoid time lapses longer than 2 min for two consecutive time points.
- Define the time settings. Go to the Time menu. Click Minimize. Select Acquire until stopped.
- Define the line average. Select Seq 1. Select Line Average 2. Select Seq 2. Select Line Average 2.
- Acquire corresponding images.
- Select Format (1024 x 1024) and Speed (400). Press Start.
- Press Stop after the desired image acquisition time.
- Save the file. Go to Project. Right click on the corresponding file. Press Save as.
- Name the file appropriately. Select the adequate folder. Press Save.
- Euthanize the animal (cervical dislocation) and collect tissues for ulterior analysis, if needed.
3. Image analysis: Determine cell shedding rate and the intestinal epithelium
- Cell shedding rate
- Launch the image acquisition software.
- Go to Open projects. Select the appropriate file.
- Define the total time of image acquisition.
- Select the last complete z stack. Select the last Z position from the previously selected time point. Read the time on the right bottom corner of the screen (total time of image acquisition).
- Select a villus.
- Select the appropriate Z stack position (surface of the villus, but deep enough to avoid the interference with already shed cells and other components present at the lumen).
- Measure the length of basal membrane. Select the ruler tool. Divide the villus in several lines to cover or draw the whole villus perimeter. Add the different values (total length of the basal membrane). Delete the segments of the Ruler tool.
- Count the number of events occurring during the whole duration of the image acquisition.
- Divide the villus into segments with an adequate size. Analyze the sequence of events/segment by sliding the bar through the different time points. Annotate the identified cell shedding events.
- Calculate the number of shedding events/time/length of basal membrane, which indicates the cell shedding rate. Count up to 5 villi/video, and at least two videos/mouse. Calculate the mean of these 10 measurements.
- Leakage
- Select a villus.
- Select the appropriate Z stack position (surface of the villus, but deep enough to avoid the interference with already shed cells and other components present at the lumen). Count total number of epithelial cells/villus.
- Count the number of leakage points (Para-cellular presence of rhodamine dextran. This event is transitory, which can be confirmed by checking previous and ulterior acquired pictures).
- Calculate the number of leakage/total number of epithelial cells. Analyze 10 different villus/sample/video and calculate the average number of leakage and permeable cells/villus.
Representative Results
The protocol presented here describes an intravital microscopy-based approach to visualize intestinal epithelial leakage and observe cell shedding performance in the gut in real-time. Briefly, mice are anesthetized and submitted to surgical preparation in order to expose the surface of the small intestine mucosa. IECs are then stained via topical application of acriflavine; while luminal rhodamine B-dextran is used as tracers to detect transmucosal passage from the lumen to the sub-epithelial space. Thus, the surgical preparation and the anesthetized mouse are placed on a slide mounted in a Petri dish and imaged over time using CLSM (Figure 1). Post-acquisition analysis permits the calculation of epithelial cell shedding rate (number of cell shedding events/minute/length of basal membrane) as well as the percentage of transitory leakage (paracellular permeability) and "permeable cells" (transcellular permeability) at a determined time point.
In order to induce alterations of epithelial integrity, we took advantage of the previously described IEC-specific GGTase-deficient conditional mouse model (Pggt1biΔIEC mice), generated via the LoxP-Cre system24. As described before, Pggt1biΔIEC mice (Figure 2A) developed severe intestinal pathology as shown by increased histological damage score in small intestine (Figure 2B). Increased intestinal epithelial permeability could be detected via tracer in vivo experiments using orally administered FITC-dextran (4 kDa) (Figure 2C), and then confirmed via intravital microscopy (Figure 2D-2E). While rhodamine dextran is restricted to the luminal compartment in control mice, we could detect the tracer within the sub-epithelial compartment upon abrogation of GGTase expression within IECs in Pggt1biΔIEC mice. During image acquisition, cell shedding events could be identified as cells moving out of the epithelial monolayer into the lumen, leading to temporary gaps in the sealing of the epithelium, which are finally closed by the contact between neighboring cells, so called zip-effect (Figure 3A). We could clearly observe these gaps, what we call temporary epithelial leakage both in control and Pggt1biΔIEC mice, although the frequency of these phenomenon was higher in the latter (Figure 3B,3C). Interestingly, we could also identify other cells where dextran could be detected intracellularly, so called "permeable cells"; these events occurred mainly in Pggt1biΔIEC mice (Figure 3B,3D). Together, taking advantage of the here presented intravital microscopy approach, we could determine that impaired epithelial integrity in Pggt1biΔIEC mice leads to cell shedding performance alterations and increased para- and trans-cellular epithelial permeability in the gut.
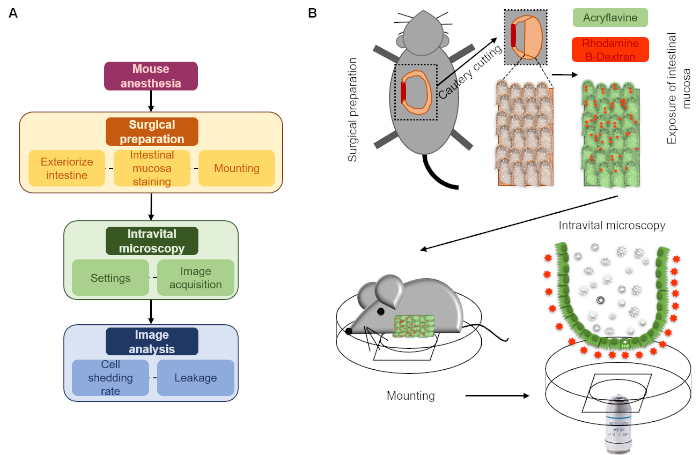
Figure 1: Schematic description of the here presented intravital microscopy approach. (A) Flow chart. (B) Diagram. After surgical preparation, intestinal mucosa topically stained with acriflavine and rhodamine dextran is mounted on a cover-slide embedded on a Petri dish to allow perfusion with a saline solution (luminal surface down). The intestinal mucosa is then imaged using a CLSM microscope over time. Please click here to view a larger version of this figure.

Figure 2: Impaired epithelial integrity in Pggt1biΔIEC mice leading to increased intestinal permeability. (A) Western blot showing tamoxifen-induced abolished GGTase-1B expression within IECs in Pggt1biΔIEC versus control mice. (B) Histological score of the small intestine from the control and Pggt1biΔIEC mice. (C) Quantification of intestinal epithelial permeability in vivo measured by transmucosal passage of orally administered FITC-Dextran (4 kDa). (D) Diagram describing the direction of image acquisition, from the lumen downwards to the villus axis. Representative pictures from a z-stack. (E) Representative pictures of intravital microscopy using topically applied acriflavine and rhodamine B-dextran from control and Pggt1biΔIEC mice, as described in this manuscript. Data are expressed as Mean ± SEM. Non-paired t-test. * P value ≤ 0.05; *** P value ≤ 0.0001. Please click here to view a larger version of this figure.
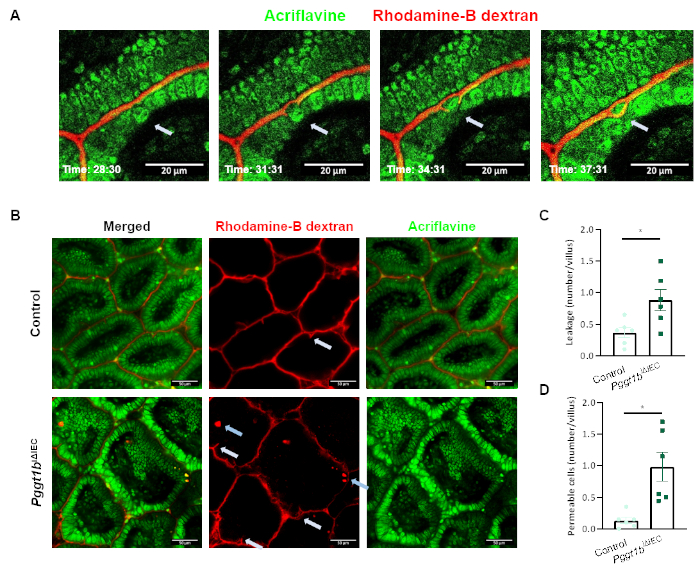
Figure 3: Epithelial cell shedding performance and para/trans-cellular epithelial permeability using intravital microscopy. (A) Representative pictures showing a cell shedding event at real-time (white arrow). The shed cell is extruded from the epithelial monolayer to the lumen. Neighboring cells seal the temporary leakage (zip-effect) to avoid loss of barrier function. (B) Representative picture showing leakage (white arrows) and permeable intestinal epithelial cells (blue arrows). (C-D) Quantification of temporary leakage (C) and permeable cells (D) in control and Pggt1biΔIEC mice. Data are expressed as Mean ± SEM. Non-paired t-test. * P value ≤ 0.05; *** P value ≤ 0.0001.Please click here to view a larger version of this figure.
Discussion
Although technically challenging, intravital microscopy-based methodology represents a unique experimental approach to visualize highly dynamic cellular process in real time, such as cell shedding performance. Thus far, there is no alternative experimental approach to visualize cell extrusion in vivo. We believe that this protocol can contribute to the description of diverse cellular processes playing a role in the maintenance of intestinal homeostasis.
Taking advantage of intravital microscopy, the method presented here enables real-time visualization of intestinal epithelial cell shedding in living animals. Therefore, topically stained intestinal mucosa (acriflavine and rhodamine B-dextran) of anesthetized mice is imaged up to single-cell resolution using confocal microscopy. In combination with standard tracer experiments, it permits the identification of intestinal permeability disturbances in vivo and the distinction between para- and trans-cellular permeability. In combination with reporter mice, similar protocols exploited to monitor epithelial cell shedding could show tight junction redistribution to seal the transitory leakage left by the extruded cells (zip-like effect)22,26.
Besides epithelial cell shedding, modifications on the method presented here have been adapted to the analysis of other highly dynamic cellular processes occurring at the surface of the intestinal mucosa. For instance, intravenous administration of tracer molecules and blood vessel staining with anti-CD31 antibodies provided the opportunity to evaluate endothelial integrity in the gut27. Beyond the epithelium, similar approaches have been used to investigate gut homing properties of immune cells28,29, as well as interaction between immune cells and IECs in the context of intestinal infection23.
Despite the plethora of potential applications of the here described protocol, there also exist some limitations to its use. Light scattering within the specimen impairs the acquisition of imaging deep inside the tissue. In the case of small intestine, the image acquisition is confined to phenomena occurring at the surface of the mucosa (villus tip). On the other hand, the structure and function of the gut itself limits the performance of intravital microscopy. Despite optimal organ optical properties, peristaltic tissue contraction as well as flow-induced movement of the intestinal villi imply frequent modifications on the focus plane during image acquisition.
Potential modifications on the here presented protocol might overcome some of these limitations. The use of alternative microscopy techniques, such as one-photon or multiphoton microscopy, implies higher penetrance depth in order to visualize focus planes located up to 50-100 µm below the surface of the sample. However, it is important to consider the compromise between image resolution and acquisition time, since these experiments aim at the observation of fast and dynamic processes. In terms of microscope configuration and/or settings, the use of longer wavelengths as well as the selection of long free working distance objectives with high numerical aperture entail optimized settings for intravital imaging.
Together, optimal experimental design taking into account the selection of appropriate fluorescent dyes and the microscopy technique, as well as the configuration of the imaging device should be considered as key steps in order to obtain a successful outcome from these experiments. As future perspective, the development of image-based prediction/quantification tools might facilitate the interpretation of acquired data.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The research leading to these results has received funding from the People Program (Marie Curie Actions) under REA grant agreement number 302170 of the European Union´s Seventh Framework Programme (FP7/2007-2013); the Interdisciplinary Center for Clinical Research (IZKF) of the University Erlangen-Nuremberg; the Collaborative Research Center TRR241 and the Clinical Research Group KFO257 of the German Research Council (DFG); and the DFG.
Materials
| Acriflavine hydrochloride | Sigma Aldrich | A8251 | 1 mg/mL solution in PBS |
| Deltaphase isothermal pad | BrainTree | B-DP-PAD | – |
| Gemini Cautery System | BrainTree | B-GEM-5917 | – |
| Ketamin | WDT | 9089.01.00 | |
| LAS X | Leica | – | – |
| LSM microscope SP8 | Leica | – | – |
| PBS | Biochrom | L182 | |
| Rhodamine B dextran | Invitrogen | D1824 | 10,000 kDa MW; 2 mg/mL solution |
| Standard forceps (Dumont SS) | Fine Science Tools | 11203-23 | – |
| Straight fine scissors | Fine Science Tools | 14060-10 | – |
| Tamoxifen | Sigma Aldrich | T5648 | 50 mg/mL in ethanol |
| Xylazin | Bayer | 1320422 |
Referenzen
- Buhner, S., et al. Genetic basis for increased intestinal permeability in families with Crohn’s disease: role of CARD15 3020insC mutation?. Gut. 55 (3), 342-347 (2006).
- Pastorelli, L., De Salvo, C., Mercado, J. R., Vecchi, M., Pizarro, T. T. Central Role of the Gut Epithelial Barrier in the Pathogenesis of Chronic Intestinal Inflammation: Lessons Learned from Animal Models and Human Genetics. Frontiers in Immunology. 4, 280 (2013).
- Kiesslich, R., et al. Local barrier dysfunction identified by confocal laser endomicroscopy predicts relapse in inflammatory bowel disease. Gut. 61 (8), 1146-1153 (2012).
- Dourmashkin, R. R., et al. Epithelial patchy necrosis in Crohn’s disease. Human Pathology. 14 (7), 643-648 (1983).
- Soderholm, J. D., et al. Augmented increase in tight junction permeability by luminal stimuli in the non-inflamed ileum of Crohn’s disease. Gut. 50 (3), 307-313 (2002).
- Wittkopf, N., Neurath, M. F., Becker, C. Immune-epithelial crosstalk at the intestinal surface. American Journal of Gastroenterology. 49 (3), 375-387 (2014).
- van der Flier, L. G., Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annual Review of Physiology. 71, 241-260 (2009).
- Fan, Y., Bergmann, A. Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell!. Trends in Cell Biology. 18 (10), 467-473 (2008).
- Ryoo, H. D., Gorenc, T., Steller, H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Developmental Cell. 7 (4), 491-501 (2004).
- Rosenblatt, J., Raff, M. C., Cramer, L. P. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Current Biology. 11 (23), 1847-1857 (2001).
- Coste, A., Oktay, M. H., Condeelis, J. S., Entenberg, D. Intravital Imaging Techniques for Biomedical and Clinical Research. Cytometry A. , (2019).
- Sorg, H., Krueger, C., Vollmar, B. Intravital insights in skin wound healing using the mouse dorsal skin fold chamber. Journal of Anatomy. 211 (6), 810-818 (2007).
- Ritsma, L., et al. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Science Translational Medicine. 4 (158), 158ra145 (2012).
- Nakasone, E. S., et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 21 (4), 488-503 (2012).
- Sipkins, D. A., et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 435 (7044), 969-973 (2005).
- Kienast, Y., et al. Real-time imaging reveals the single steps of brain metastasis formation. Nature Medicine. 16 (1), 116-122 (2010).
- Nguyen, V. X., Nguyen, C. C., De Petris, G., Sharma, V. K., Das, A. Confocal endomicroscopy (CEM) improves efficiency of Barrett surveillance. Journal of Interventional Gastroenterology. 2 (2), 61-65 (2012).
- Kiesslich, R., Goetz, M., Neurath, M. F. Confocal laser endomicroscopy for gastrointestinal diseases. Gastrointestinal Endoscopy Clinics of North America. 18 (3), 451-466 (2008).
- Kiesslich, R., et al. Chromoscopy-guided endomicroscopy increases the diagnostic yield of intraepithelial neoplasia in ulcerative colitis. Gastroenterology. 132 (3), 874-882 (2007).
- Krishnamurthy, S., et al. Confocal Fluorescence Microscopy Platform Suitable for Rapid Evaluation of Small Fragments of Tissue in Surgical Pathology Practice. Archives of Pathology & Laboratory Medicine. 143 (3), 305-313 (2019).
- Li, B. R., et al. In vitro and In vivo Approaches to Determine Intestinal Epithelial Cell Permeability. Journal of Visualized Experiments. (140), (2018).
- Marchiando, A. M., et al. The epithelial barrier is maintained by in vivo tight junction expansion during pathologic intestinal epithelial shedding. Gastroenterology. 140 (4), 1208-1218 (2011).
- Hoytema van Konijnenburg, D. P., et al. Intestinal Epithelial and Intraepithelial T Cell Crosstalk Mediates a Dynamic Response to Infection. Cell. 171 (4), 783-794 (2017).
- Lopez-Posadas, R., et al. Rho-A prenylation and signaling link epithelial homeostasis to intestinal inflammation. Journal of Clinical Investigation. 126 (2), 611-626 (2016).
- Duckworth, C. A., Watson, A. J. Analysis of epithelial cell shedding and gaps in the intestinal epithelium. Methods in Molecular Biology. 763, 105-114 (2011).
- Watson, A. J., et al. Epithelial barrier function in vivo is sustained despite gaps in epithelial layers. Gastroenterology. 129 (3), 902-912 (2005).
- Haep, L., et al. Interferon Gamma Counteracts the Angiogenic Switch and Induces Vascular Permeability in Dextran Sulfate Sodium Colitis in Mice. Inflammatory Bowel Diseases. 21 (10), 2360-2371 (2015).
- Lopez-Posadas, R., et al. Inhibiting PGGT1B Disrupts Function of RHOA, Resulting in T-cell Expression of Integrin alpha4beta7 and Development of Colitis in Mice. Gastroenterology. , (2019).
- Fischer, A., et al. Differential effects of alpha4beta7 and GPR15 on homing of effector and regulatory T cells from patients with UC to the inflamed gut in vivo. Gut. 65 (10), 1642-1664 (2016).

