Real-time Analysis of Transcription Factor Binding, Transcription, Translation, and Turnover to Display Global Events During Cellular Activation
Summary
This protocol describes the combinatorial use of ChIP-seq, 4sU-seq, total RNA-seq, and ribosome profiling for cell lines and primary cells. It enables tracking changes in transcription-factor binding, de novo transcription, RNA processing, turnover and translation over time, and displaying the overall course of events in activated and/or rapidly changing cells.
Abstract
Upon activation, cells rapidly change their functional programs and, thereby, their gene expression profile. Massive changes in gene expression occur, for example, during cellular differentiation, morphogenesis, and functional stimulation (such as activation of immune cells), or after exposure to drugs and other factors from the local environment. Depending on the stimulus and cell type, these changes occur rapidly and at any possible level of gene regulation. Displaying all molecular processes of a responding cell to a certain type of stimulus/drug is one of the hardest tasks in molecular biology. Here, we describe a protocol that enables the simultaneous analysis of multiple layers of gene regulation. We compare, in particular, transcription factor binding (Chromatin-immunoprecipitation-sequencing (ChIP-seq)), de novo transcription (4-thiouridine-sequencing (4sU-seq)), mRNA processing, and turnover as well as translation (ribosome profiling). By combining these methods, it is possible to display a detailed and genome-wide course of action.
Sequencing newly transcribed RNA is especially recommended when analyzing rapidly adapting or changing systems, since this depicts the transcriptional activity of all genes during the time of 4sU exposure (irrespective of whether they are up- or downregulated). The combinatorial use of total RNA-seq and ribosome profiling additionally allows the calculation of RNA turnover and translation rates. Bioinformatic analysis of high-throughput sequencing results allows for many means for analysis and interpretation of the data. The generated data also enables tracking co-transcriptional and alternative splicing, just to mention a few possible outcomes.
The combined approach described here can be applied for different model organisms or cell types, including primary cells. Furthermore, we provide detailed protocols for each method used, including quality controls, and discuss potential problems and pitfalls.
Introduction
In recent years, RNA-sequencing (RNA-seq) has become the standard tool to analyze all expressed RNAs within a cell or an organism1. However, to understand the whole process of cells adapting in response to a specific stimulus/drug, it is necessary to fully determine all underlying processes, ranging from mRNA transcription to processing, turnover, and translation. Short-term changes in RNA transcription can hardly be measured by total RNAseq, since changes of total RNA depend on factors e.g. RNA half-lives and transcriptional activity, which are a poor template for reflecting the adaptation of cells to environmental effects2,3. Indeed, a variety of new sequencing techniques have been developed that allow an analysis of the different steps in the process of gene regulation4 when combined in the right manner. This protocol describes how to combine some rather easily applicable sequencing techniques that allow tracking the regulation of the essential layers of mRNA in a comparative manner. For analyzing transcriptional activity, a variety of methods have been described, such as Cap-analysis of gene expression (CAGE)5, native elongation transcript sequencing (NET-seq)6, and genome-wide nuclear run-on (GRO-seq)7,8, as well as bromouridine-sequencing (Bru-seq) and 4-thiouridine-sequencing (4sU-seq), which use metabolites that are incorporated in newly transcribed RNA9,10, just to mention a few. While CAGE identifies the exact transcription start site, NET-seq and GRO-seq deliver more accurate information about reading directions, and 4sU-seq (which is the method described here) detects only newly transcribed RNA. However, 4sU-seq is highly sensitive and can be applied in different time frames to measure quantitative transcriptional activity in actively changing cells, as well as quantitative changes in mRNA processing (which happens within minutes)9,11,12. Furthermore, 4sU-seq is ideal to combine with RNA-seq to calculate RNA turnover rates for genes9. The transcription of mRNA is carried out by RNA Polymerase II (RNAPII), which is in turn influenced by a multitude of factors, such as transcription factors, histone modifications, and general activators/repressors that can even be part of the transcriptional complex. To test how many gene/promoter/enhancer regions are bound by one factor, ChIP-seq has been developed, which is now the standard method for this purpose, due to many commercially available antibodies13. However, although ChIPseq gives clear information on where regulating factors bind, it does not reflect if it indeed leads to changes in transcription14. Therefore, performing ChIP-seq with 4sU-seq is the ideal combination for such biological questions. Regulation of gene expression can also occur at a later stage, since mRNA and protein levels do not necessarily correlate15,16, indicating potentially significant regulation on the translational or post-translational level, depending on the context. In the year 2011, ribosome profiling had been first combined with RNA-seq and is now the method of choice to quantify changes that occur rapidly in protein, since there are still some sensitivity limits with mass spectrometry17. Indeed, translation rates obtained from such methods have been shown to deliver a relatively good estimate of changes in protein levels (at least measured for long-term changes) and allow an even more detailed view on the process of translation, e.g. the determination of start-side and alternative translation frames17. The combinatorial use of all four methods can be used in steady state, between various cell types, or in a time-serial experiment of a rapidly changing cell11. This use provides a genome-wide overview of changes in transcription factor binding influencing RNA transcription, processing, and translation.
Protocol
All methods are in accordance and compliance with the Helmholtz Zentrum München institutional, state, and federal guidelines.
1. Preparation
- Make a detailed plan of the experimental setup including a time schedule, when to add 4sU to the cell culture medium, and when to harvest the cells for each method. Depending on the biological question, carefully consider time points for sample drawing, 4sU-labeling time, and concentration (Table 1).
NOTE: Verify the impact of 4sU on cell viability and stress response in advance (see "Verify optimal 4sU-labeling conditions" in Representative Results and Figure 1). It is recommended to perform a preliminary test of each method with at least one sample. Verify if the quality and amount of RNA/DNA is sufficient for deep sequencing (see dedicated parts of the protocol), and practice fast but gentle handling of the cells during the experiment. - Calculate the number of cells needed for every time point of each method (see Table 2 for a rough estimation when using primary T cells). Also, consider sequencing fewer samples of total RNA than just that of 4sU RNA (e.g., just for time point translation rates or RNA turnover rates should be calculated). To confirm and verify the significance of the results (recommended), create at least one biological replicate.
NOTE: Importantly, for all methods and consecutive time points, samples must be from the same starting pool of cells. At least one dedicated researcher for each method is recommended. - Prepare everything necessary in advance (e.g., aliquoted 4sU, cycloheximide, Mammalian Lysis Buffer, and 1% formaldehyde). After addition of 4sU, avoid exposing the cells to bright light, as this may lead to crosslinking of 4sU-labeled RNA to cellular proteins18.
- Pool all cells of interest in one flask just prior to treatment (Figure 2). Count the cells (e.g., with a hemocytometer) and use the required amount for the untreated control of each method (do not forget to also label the untreated control for 4sU-seq with 4sU). Treat remaining cells and immediately split the required number of cells for each time point and method. Handle cells as quickly as possible to minimize stress due to changes in temperature or CO2 levels.
NOTE: For example, samples are taken 1 h, 2 h, and 3 h after treatment, then one quarter of the cells is used for untreated control, and three quarters are used for treated control.
2. 4sU-Labeling
NOTE: This protocol is modified from Rädle, et al.19 Refer to their protocol for further information regarding metabolic labeling with 4sU. For all methods and consecutive time points, samples must originate from the same starting pool of cells.
- Start of Labeling
- Thaw aliquoted 4sU just before use. Add 4sU at each time point directly to the medium containing cells of interest (refer to Table 2 for recommended T cell numbers, least 60 µg RNA per time point), mix gently, and place back into the incubator. Dispose remaining 4sU (do not refreeze).
- At the end of labeling, collect cells (e.g., cell scraper) and centrifuge at 330 x g for 5 min at 4 °C in polypropylene tubes (which resist high g forces). Aspirate medium and add reagent for RNA isolation (≥1 mL per 3 x 106 cells, see materials for recommendation which to use) to each tube. Fully resuspend the pellet (≥1 mL per 3 x 106 cells), incubate for 5 min at room temperature (RT), and freeze samples at -20 °C. Samples can be stored at -20 °C for at least 1 month.
Caution: The reagents used for RNA isolation are extremely hazardous when getting in contact with skin or eyes. Handle these with care and consider the safety instructions.
- RNA preparation using modified RNA isolation protocol
- Add 0.2 mL chloroform per 1 mL reagent for RNA isolation, and mix thoroughly by shaking for 15 s. Proceed as mentioned in the metabolic labeling protocol (step 1 – 12, 2. RNA preparation using modified trizol protocol) from Rädle et al.19
- Measure RNA concentration (see Table of Materials), according to the manufacturer's instructions. Use this RNA for total RNA-seq as well (see step 3. Total RNA-seq) or store at -80 °C for at least 1 month.
- Thiol-Specific Biotinylation of Newly Transcribed RNA
- Start with 30 – 80 µg of total cellular RNA. 60 µg RNA should yield sufficient amounts of newly transcribed RNA.
- Prepare labeling reaction. Pipette in the following order (per µg RNA): 1 µL 10x Biotinylation Buffer, 7 µL RNA (containing 1 µg RNA diluted in nuclease-free H2O), and 2 µL biotin-HPDP (1 mg/mL). Add biotin-HPDP last and mix immediately by pipetting. Wrap tubes with aluminum foil to avoid exposure to bright light. See the discussion for an alternative to biotin-HPDP. Incubate at RT for 1.5 h with rotation.
- Centrifuge appropriate 2 mL tubes (see Table of Materials) at 15,000 x g for 2 min. Pipette all of the biotinylated RNA into the pre-spun 2 mL tube, add an equal volume of chloroform, and mix vigorously. Incubate for 2 – 3 min until the phases begin to separate and bubbles start to disappear.
- Centrifuge at 15,000 x g for 15 min at 4 °C. Carefully transfer the upper aqueous phase into a new tube.
- Repeat steps 2.3.3. and 2.3.4. once. Add 10% of the volume of NaCl (5 M) and an equal volume of isopropanol to the aqueous phase. Centrifuge at 20,000 x g for 20 min at 4 °C. Discard supernatant.
- Add an equal volume freshly prepared 75% ethanol. Centrifuge at 20,000 x g. Discard the supernatant, spin briefly, and remove the remaining ethanol. Fully resuspend in 30 – 100 µL H2O (use 1 µL H2O per 1 µg input RNA from step 2.3.1) by pipette mixing.
- Verify RNA integrity by electrophoretic analysis, or take an aliquot and verify later.
- Separation of Newly Transcribed (Labeled) and Preexisting (Unlabeled) RNA
- Remove paramagnetic beads (see Table of Materials) from 4 °C storage and let it stand for at least 30 min to bring them to room temperature. Heat 4sU washing buffer (3 mL per sample) to 65 °C.
- Prepare 100 mM dithiothreitol (DTT) solution. Weigh DTT on an ultra-fine scale and add the required amount of nuclease-free water. Always prepare fresh. Use 200 µL per sample.
- Heat biotinylated RNA samples (1 µg/µL) to 65 °C for 10 min to denature and immediately place on ice. Add 100 µL streptavidin beads to biotinylated RNA and incubate at RT for 15 min with rotation.
- Place an appropriate column (see Table of Materials for recommendations) for each sample into the magnetic stand, and pre-equilibrate each column with 1 mL room temperature 4sU washing buffer.
NOTE: This will take approximately 5 – 10 min. If any of the columns do not initiate draining after 5 min, press gently on top of the column with a gloved finger. - Apply an RNA/beads mix to the middle of each column. Discard the flow-through, unless unlabeled RNA needs to be recovered. If so, collect the flow-through and at least the first wash. Perform the recovery of RNA as described by Rädle et al. (step 1 – 7, 7. Recovery of Unlabeled, Unbound RNA)19.
- Wash three times with 0.9 mL 4sU washing buffer (preheated to 65 °C from step 2.4.2.) and 0.9 mL RT 4sU washing buffer, respectively.
- Use paramagnetic beads to recover newly transcribed RNA. Pipette 400 µL of well-dispersed RT paramagnetic beads in a tube per sample and place underneath each column. Elute newly transcribed RNA with 100 µL 100 mM DTT. Wait for 3 min and perform a second elution with 100 µL 100 mM DTT. (Optional: Perform elution and recovery as described by Rädle et al.)19
- Mix newly transcribed RNA/beads thoroughly by pipette mixing 10 times and proceed according to the manufacturer's guideline. Elute RNA in 11 µL nuclease-free H2O.Quantify RNA using a suitable fluorometer (see Table of Materials). RNA can be stored at -80 °C for at least 1 month.
NOTE: Newly transcribed RNA can be used to prepare cDNA libraries for next-generation sequencing (see Table of Materials for a suggestion on which kit to use) or further downstream analyses. 100 – 500 ng RNA are sufficient for most library preparation kits (see Discussion).
3. Total RNA-Seq
- Directly take RNA from 4sU labeled RNA after RNA preparation using the modified reagent for RNA isolation protocol for total RNA-seq (see step 2.2.2.)
- For library preparation, dilute an RNA aliquot to a final concentration of 50 – 100 ng/µL. Use the same kit as for library preparation of newly transcribed RNA. 100 – 500 ng RNA are sufficient for most library preparation kits.
4. Ribosome Profiling
NOTE: For all methods and consecutive time points, samples must be from the same starting pool of cells . For recommendations on which kit to use, refer to the Table of Materials.
- Preparation and Isolation of Ribosome Protected Fragments (RPFs):
- Use appropriate amounts of cells for each time point (refer to Table 2 for recommended T cell numbers). Treat adherent cells with cycloheximide as described in the manufacturer's protocol.
Caution: Cycloheximide is highly toxic and may cause mutations. Avoid skin contact and inhalation. - Collect and pool non- or semi-adherent cells from each time point into one polypropylene tube and adjust the final concentration to 1 x 106 cells per mL of cell-specific medium (e.g., supplemented RPMI for T cells). Add cycloheximide with a final concentration of 0.1 mg/mL, mix by inverting the polypropylene tube, and incubate for 1 min. Centrifuge cells for 5 min at 330 x g at 4 °C. Aspirate medium and wash cells with at least 10 mL PBS supplemented with cycloheximide (final concentration of 0.1 mg/mL).
- Centrifuge cells for 5 min at 330 x g at 4 °C. Aspirate medium and add 100 µL mammalian cell lysis buffer per 10 x 106 cells. Mix by pipetting and expel through a sterile 22 25-gauge needle to lyse the cells completely.
- Transfer the cell lysate to a precooled 1.5 mL tube. Incubate for 10 min on ice with periodic inversions. Centrifuge for 10 min at 20,000 x g at 4 °C to clarify the lysate. Transfer the supernatant to a precooled 1.5 mL tube.
- Prepare a 1:10 dilution of the lysate with nuclease-free water and record an A260reading using a spectrophotometer. Use nuclease-free water as a blank and a 1:10 dilution of mammalian cell lysis buffer as the standard. Calculate the A260/mL concentration of the lysate according to the following equation:
(A260 cell lysate – A260 Mammalian Lysis Buffer) x 10 dilution factor = A260/mL - Create 200 µL aliquots of the lysate on ice and proceed with nuclease treatment.
NOTE: Optionally, prepare a 100 µL aliquot for total RNA, add 10 µL of 10% SDS and mix. Store at 4 °C and proceed with 4.3.2. Using total RNA from 4sU-labeled RNA is recommended (see step 3. Total RNA-seq).
- Use appropriate amounts of cells for each time point (refer to Table 2 for recommended T cell numbers). Treat adherent cells with cycloheximide as described in the manufacturer's protocol.
- Ribosome footprinting
- Perform nuclease treatment immediately without freezing the lysate. Add 7.5 units of nuclease (included in the recommended kit) for each A260 of lysate. For example: 80 A260/mL lysate x 0.2 mL lysate x 7.5 U/A260nuclease = 120 U nuclease.
NOTE: Optionally, titrate the nuclease for digestion as described by the manufacturer. - Incubate the nuclease reaction for 45 min at RT with gentle mixing. Freeze 200 µL aliquots of the lysate with liquid nitrogen and store at -80 °C, or stop the nuclease reaction by adding 15 µL RNase inhibitor to each 200 µL aliquot, and continue with step 4.3.2.
- Perform nuclease treatment immediately without freezing the lysate. Add 7.5 units of nuclease (included in the recommended kit) for each A260 of lysate. For example: 80 A260/mL lysate x 0.2 mL lysate x 7.5 U/A260nuclease = 120 U nuclease.
- Purification of RPFs
- Unfreeze one sample of nuclease digested RPFs and add 15 µL RNase inhibitor. Keep the samples on ice.
- Purify the RPFs according to the manufacturer's protocol (column purification is recommended) and measure RNA concentration on a spectrophotometer.
- rRNA Depletion
- Use 5 µg of purified RPFs for rRNA depletion.
- Follow the manufacturer's protocol (step 1 – 2, Primary rRNA depletion) for rRNA depletion. Measure RNA concentration of rRNA depleted RPFs on a spectrophotometer.
- PAGE Purification of RPFs
- Use 500 ng of rRNA depleted RPFs for PAGE purification.
- Prepare RNA Control, samples, and ladder for PAGE purification. Mix 5 µL RNA Control and 5 µL denaturing gel loading dye in a 0.5 mL microcentrifuge tube. Mix 10 µL of each RPF with 10 µL of denaturing gel loading, respectively. Prepare a ladder aliquot (4 µL 20/100 ladder, 1 µL nuclease-free water, and 5 µL denaturing gel loading dye). Load it between each sample and control to prevent crosscontamination.
- Denature samples and ladder by incubating at 95 °C for 5 min and immediately place on ice. Load 20 µL of each sample (optionally, load 10 µL and freeze remaining samples at 20 °C) separated by 10 µL of prepared ladder onto a 12% or 15% urea-polyacrylamide gel. Load 10 µL of RNA Control. Run the gel until the bromophenol blue band reaches the bottom of the gel (180 V, ~70 min) (Figure 3).
- Stain the gel according to the manufacturer's protocol at 4 °C. Use a dark-field transilluminator that emits blue light to visualize the RNA. Excise gel slices for each sample corresponding to ~28 and 30 nt in length. Take RNA Control as reference and excise it.
NOTE: RPFs are hardly visible. Excise slices at the size indicated by the RNA Control – containing two oligos of 28 and 30 nt in length – even if samples are not visible. - Puncture a hole in the bottom of 0.5 mL microcentrifuge tubes with a sterile 20-gauge needle. Transfer each gel slice into a separate tube and place capped tubes in a 1.5 mL tube. Centrifuge for 2 min at 12,000 x g. Repeat centrifugation if gel slices are not completely shred into the 1.5 mL tube.
- Elute the RNA from disrupted gel slices with 400 µL nuclease-free water, 40 µL ammonium acetate (5 M), and 2 µL SDS (10%) each overnight at 4 °C.
- Transfer the slurry to 1.5 mL filter tubes (provided with the recommended kit) with a 1 mL pipette tip (wide-bore tip or self-made 1 mL tip with cut end). Centrifuge for 3 min at 2,000 x g to separate eluted RNA from gel slices. Gently pipette aqueous solution into a 1.5 mL tube. Add 2 µL glycogen (provided with the recommended kit) and 700 µL 100% isopropanol and store at -20 °C for at least 1 h.
- Centrifuge at 4 °C for 20 min at 13,000 x g. Discard supernatant. Wash the pellet with pre-chilled freshly prepared 80% ethanol at 4 °C for 10 min at 13,000 x g. Discard supernatant and air-dry. Resuspend each sample in 20 µL and the RNA Control in 8 µL nuclease-free water. Store at -20 °C if needed.
- Fragmentation, End Repair, 3' Adaptor Ligation, Reverse Transcription
- Perform the procedure as described by the manufacturer's protocol (Fragmentation and end repair, 3' adapter ligation, and reverse transcription).
- PAGE Purification of cDNA
- Prepare samples, RNA Control and ladder for PAGE purification: Mix 10 µL of each sample and RNA Control with 10 µL denaturing gel loading dye, respectively. Prepare a ladder aliquot (4 µL 20/100 ladder, 1 µL nuclease-free water, 5 µL denaturing gel loading dye). Load it between each sample and control to prevent crosscontamination.
- Denature samples and ladder by incubating at 95 °C for 5 min and immediately place on ice. Load 20 µL of each sample (Optionally, load 10 µL and freeze remaining samples at 20 °C) separated by 10 µL of prepared ladder onto a 10% polyacrylamide/7 – 8 M urea/TBE gel. Load 10 µL of RNA Control. Run the gel until the bromophenol blue completely migrates out of the gel (180 V, ~60 min).
- Stain the gel according to the manufacturer's protocol at 4 °C. Use a dark-field transilluminator that emits blue light to visualize the RNA and excise the gel slices for each sample corresponding to ~70 – 80 nt.
- Proceed as described in step 4.5.5 – 4.5.8 and resuspend each sample in 10 µL nuclease-free water.
- cDNA Circularization
- Prepare enough circularization master mix for all reactions by combining the following reagents for each sample on ice: 4.0 µL Circularization Reaction Mix, 2.0 µL ATP, 2.0 µL MnCl2, and 2.0 µL Ligase.
- Add 10 µL of the master mix to each sample. Mix gently and centrifuge briefly. Incubate samples at 60 °C for 2 h. Immediately place samples on ice.
- PCR Amplification
- Follow the manufacturer's protocol (step 1 – 3, PCR amplification) for PCR amplification. Use 4 µL of circularized cDNA for amplification with 9 PCR cycles for primary T cells, to achieve best results.
- Purify libraries and check their size distribution, according to the manufacturer's protocol (step 4 – 8, PCR amplification). The expected size of the amplified library is 140 – 160 bp (see Figure 4).
- For sequencing libraries, refer to the manufacturer's protocol and the sequencing facility for further guidance.
5. ChIP-Seq
NOTE: This protocol is modified from Blecher-Gonen, et al.14 Refer to their protocol for further information regarding ChIP-seq. For all methods and consecutive time points, samples must be from the same starting pool of cells.
- Crosslinking and Harvesting the Cells
- Crosslink appropriate number of cells (refer to Table 2 for recommended T cell numbers) for each time point with a final concentration of 1% formaldehyde in a cellspecific medium (e.g., supplemented RPMI for T cells) for 10 min at room temperature with gentle rocking. Stop the crosslinking reaction by the addition of glycine to a final concentration of 0.125 M.
- Centrifuge cells at 330 x g for 5 min at 4 °C. Discard the supernatant and wash the cells in ice-cold PBS. Repeat step 5.1.2 twice and freeze cell pellets at -80 °C. Frozen pellets can be stored for at least 6 months.
- Cell Lysis and Sonication
NOTE: During all cell lysis and sonication steps, samples must be kept on ice or at 4 °C to minimize crosslink reversal and protein degradation.- Resuspend cell pellets in 1 mL ice-cold cell lysis buffer with freshly added protease inhibitors to isolate the nuclei (add phosphatase inhibitors optionally). Incubate for 10 min on ice and centrifuge at 2,600 x g for 5 min at 4 °C.
- Aspirate supernatant and resuspend nuclei pellet in 1 mL ice-cold nuclei lysis buffer with freshly added protease inhibitors (optionally: add phosphatase inhibitors). Incubate for 10 min on ice. Sonicate the cells to generate a mean DNA size fraction of 0.2 – 1.0 kb (see Figure 5).
NOTE: Sonication conditions need to be optimized according to the cell type and further conditions (e.g., cell number, volume, and buffer). For primary T cells, sonication for 20 – 25 cycles is recommended (see materials for detailed description). - Take a 20 – 50 µL aliquot of the sheared chromatin and heat for 10 min at 95 °C and 1,000 rpm shaking to perform a fast reverse crosslink and verify chromatin size. Add 2 – 5 µL Proteinase K and incubate for 20 min at 56 °C and 1,000 rpm shaking. Perform heat inactivation for 10 min at 95 °C and 1,000 rpm shaking. Purify chromatin with an appropriate kit (see Table of Materials). Check chromatin size on a 1% agarose gel and use 100 bp Plus Marker.
- Centrifuge sheared chromatin with a mean DNA size fraction of 0.2 – 1.0 kb for 10 min at 20,000 x g and 4 °C to pellet insoluble chromatin and debris. Transfer the supernatant to a new tube and keep on ice.
- Keep 5 – 10% of sonicated chromatin as input. Freeze at -20 °C (used in step 5.5.2).
- Couple Antibody to Beads
- Couple 10 µg antibody (e.g., anti-RNA Pol II; anti-Histon H3K36me3) in 220 µL PBS (with 0.5% BSA and 0.5% Tween 20) to 80 µL superparamagnetic beads coupled to protein G (see Table of Materials) for at least 1 h at room temperature with rotation.
- Place the tubes on a magnet. Wait until all beads are bound to the magnet and remove the supernatant. Further block with 6 µL sonicated salmon sperm DNA in PBS (with 0.5% BSA and 0.5% Tween 20) for 30 min at room temperature with rotation.
- Place the tubes on a magnet. Wait until all beads are bound to the magnet and remove the clear supernatant. Wash beads with ChIP IP buffer three times.
- Chromatin Immunoprecipitation
- Dilute chromatin to 1 mL total volume in nuclei lysis buffer with freshly added protease inhibitors (optionally, add phosphatase inhibitors). Add ChIP IP buffer with freshly added protease inhibitors (optionally, add phosphatase inhibitors) to a final volume of 3 mL. Keep on ice or at 4 °C while antibody is coupled to the beads.
- Add diluted chromatin to the antibody coupled beads from step 5.3.3 and incubate over night at 4 °C with gentle rotation.
- Wash with the following buffers (1 mL each, see Table of Materials) at room temperature for 5 min with rotation, place tubes back on the magnet and remove supernatant: Wash Buffer I, Wash Buffer II, Wash Buffer III, and 2x TE pH 8.0 respectively.
- Discard supernatant and air-dry for ~5 min.
- Reverse crosslinking
- Remove samples from the magnet. Add 50 µL elution buffer and mix by pipetting to elute protein-DNA complexes from the beads.
- Include input sample(s) from this step forward. Add elution buffer to input sample(s) for a final volume of 50 µL (to keep the buffer composition similar to the ChIP samples) and process together with the ChIP samples.
- Mix 3 µL of elution buffer and 2 µL of RNase (DNase free). Add 5 µL of the mix to each sample and incubate for 30 min at 37 °C.
- Mix 2.5 µL proteinase K, 1 µL glycogen, and 1.5 µL elution buffer per sample. Add 5 µL of the mix to each sample (1 U proteinase K and 20 µg glycogen per sample) and incubate for 2 h at 37 °C.
- Incubate the samples at 65 °C overnight (at least 4 h) with shaking to perform reverse crosslinking.
- Place the tubes on the magnet for at least 30 s and transfer the supernatant to a new tube. Samples can be frozen at -20 °C for up to 12 months.
- DNA purification
- Add 140 µL of well-dispersed paramagnetic beads to 60 µL of sample (2.3:1 ratio). Carefully pipette up and down 25 times to mix thoroughly. Ensure that the liquid in each tube is homogenous. Incubate at room temperature for 2 min. Place the tubes on the magnet for 4 min, or until all beads are bound to the magnet, and discard the supernatant.
- Leave the tubes on the magnet and add 200 µL of freshly prepared 70% ethanol. Incubate the tubes for 30 s without disturbing the beads. Discard the supernatant and repeat this step once more. Aspirate ethanol completely and let the paramagnetic beads air-dry for 4 min.
NOTE: Incomplete ethanol removal may seriously reduce DNA recovery and yield. Dry the pellet only until it is dry. Over-drying the pellet may reduce DNA recovery and yield. - Remove the tubes from the magnet and add 20 µL 10 mM Tris-HCl (pH 8.0). Gently pipette the entire volume up and down 25 times to mix thoroughly. Incubate for 2 min at room temperature. Place the tubes back on the magnet for 4 min and transfer the supernatant to another tube.
- Measure the amount of DNA with a suitable fluorometer (see Table of Materials).
- Verify the ChIP was successful by qPCR (dilute 1 µL in 100 µl H2O and use 2 – 5 µL for qPCR). Use specific primers for a positive (known binding site of protein of interest) and negative control (e.g., a gene that is silent and/or not a target of protein of interest).
NOTE: Library preparation can be performed with 2 ng of ChIP DNA depending on the kit (see Table of Materials for a suggestion on which kit to use).
Representative Results
4sU labeling: Verify optimal 4sU-labeling conditions (apoptosis, nuclear stress, cytoplasmic stress), time, and concentration: High levels of 4sU can inhibit the production and processing of rRNA and induce cytoplasmic as well as nuclear stress30. Therefore, cells of interest should be tested for 4sU-induced stress as well as apoptosis. Western blot analysis is recommended for visualizing p53 accumulation, which indicates nuclear stress, increasing phospho-EIF2a levels which display cytoplasmic stress and fluorescence-activated cell sorting (FACS) analysis for apoptosis. High levels and lengthy exposure to 4sU or drugs like thapsigargin or arsenite can be used to induce cellular stress. To induce apoptosis or cell death, cells were treated with BH3I-1 (500 ng/µL) or incubated for 5 min at 95 °C (heat shock). Annexin V/7-AAD staining was used to determine apoptotic (Annexin V) and dead (7-AAD) cells. Labeling of in vitro generated primary Th1 cells for 0.5 h with 500 µM 4sU (final concentration) or 1 h with 200 µM 4sU neither induces signs of cellular stress nor apoptosis (Figure 1) but lead to sufficient 4sU incorporation.
RNA labeling time can also be shortened (≤5 min) which leads to an increase in short-lived intronic sequences compared to longer labeling times. To visualize co-transcriptional splicing rates, 4sU-labeling times should not exceed 30 min. For further details regarding 4sUlabeling, please refer to Rädle et al.19
Quality Control: RNA integrity is of great importance when processing RNA. It is most convenient to check the RNA quality of 4sU-labeled RNA after biotinylation by electrophoretical analysis (see Table of Materials). Consider verifying isolated RNA from step 2.2.2, especially when using it for sequencing of total RNA. RNA integrity number (RIN) should be ≥8 to ensure RNA integrity for further processing (Figure 3).
Electrophoretical analysis can also be used to verify newly transcribed RNA. Be aware that newly transcribed RNA contains significantly less mature rRNAs compared to total RNA with the typical rRNA bands being much less prominent.
Ribosome profiling: PCR amplification of the cDNA library: cDNA amplification (step 4.9.1) is a critical step to ensure good sequencing results. Analyze amplified libraries by electrophoretical analysis. A good sample of amplified libraries shows a peak around 140 – 160 bp (Figure 4A). Excessive amounts of adapter dimers should be avoided (Figure 4B) and those samples should be further purified using the PAGE purification procedure according to the manufacturer's protocol (PAGE purification of PCR products). Too much template or too many PCR cycles can result in over-amplification characterized by the appearance of higher-than-expected molecular weight bands, smeared PCR products, and adapter dimer products (Figure 4C). For most samples 1 – 5 µL of circularized cDNA and 9 PCR cycles for amplification will typically yield sufficient amounts of the correct PCR product.
ChIP: Chromatin shearing: Optimal shearing conditions need to be adjusted for each type of cell. Determine shearing conditions (e.g., number of cycles, high or low power) in advance. Use the same number of cells and the same volume for testing purposes, since a lower cell density increases the shearing efficiency. Try to avoid over- or under-shearing the chromatin. Large chromatin fragments can dramatically affect ChIP results by clogging, and over-shearing can destroy epitopes on the protein of interest, leading to a lower binding efficiency by the antibody. In this experiment, the best results were achieved when the main fraction of the sheared chromatin was around 1,000 bp or slightly lower (Figure 5A).
Verifying ChIP by qPCR: Before starting the ChIP, it is advisable to test if the used antibody is suitable for ChIP (if possible, use ChIP grade antibodies) by ChIP-qPCR. Verify the ChIP for sequencing by qPCR before starting the library preparation (see step 5.6.5). Design primers that bind to a known target site of the protein of interest. If the exact target site within a gene is unknown, several primer pairs can be used to scan the gene and associated regulatory elements. For RNAPII ChIP of Th1 cells Ifng, which is transcriptionally upregulated upon stimulation, and actin primers can be used as a positive control. Sox9 and insulin serve as a negative control, since these genes are not expressed in Th1 cells (Figure 5B). Remember not to use exon-spanning primers, which are normally used for qPCR of mRNA. An IgG control can also be used to prove specificity of the used antibody. Immunoprecipitated DNA can be measured with a suitable fluorometer (see Table of Materials). Amounts of nonspecifically bound DNA by the IgG control should be significantly lower compared to the DNA amount bound by the antibody of interest.
Replicates: proof of biological significance: It is strongly advised to perform the kinetic experiment for all methods starting from the same pool of cells to ensure cells have the same identity for all untreated and treated samples (Figure 2). Nevertheless, it is recommended to take small aliquots of main time points for each method to compare samples to a biological replicate (e.g., by qPCR, FACS analysis). This allows for a rough estimation if the treatment for both replicates was reproducible and you could proceed with sequencing. Validation of the replicates should be performed by means of bioinformatical analysis. Reproducibility of results can be assessed in terms of correlation between FPKM values between replicates and visualized using scatter plots (Figure 6).
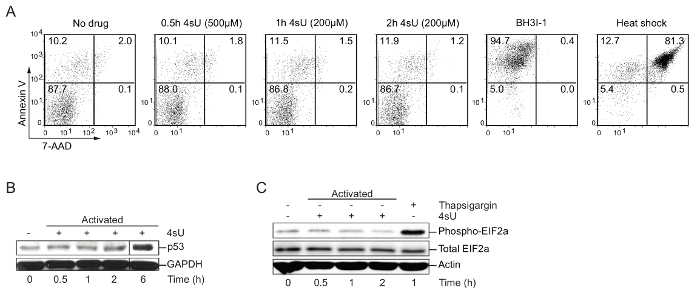
Figure 1: Verification of optimal 4sU-labeling conditions without perturbing cell physiology (Figure from Davari et al.11)
(A) Detection of cell apoptosis by FACS analysis: In vitro generated Th0 cells were treated with different concentrations of 4sU (indicated in brackets) for 0.5 h, 1 h, and 2 h, respectively. BH3I-1 treatment was used to induce apoptosis determined by Annexin V, whereas heat shock (5 min at 95 °C) was used to induce cell death determined by 7-AAD. (B) Western blot analysis for p53 of 4sU treated and activated T cells: samples were labeled with 200 µM 4sU for the indicated time of activation, except the 0.5 h time point that was labeled with 500 µM 4sU. (C) Western blot analysis of phospho-EIF2a and total EIF2a in activated Th1 cells with the same labeling conditions as in (B). Thapsigargin was used as a positive control. Please click here to view a larger version of this figure.
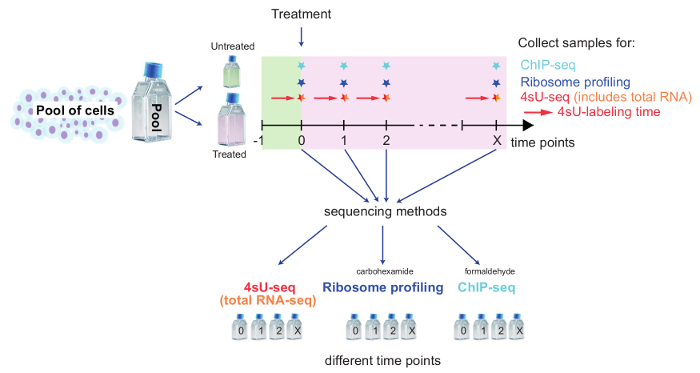
Figure 2: Schematic overview of a kinetic setup to track genome-wide changes
This scheme illustrates the setup for combining 4sU-seq, total RNA-seq, Ribosome Profiling, and ChIP-seq to study genome-wide changes upon cell treatment. Pool cells and set aside the required number of cells for untreated control. Treat remaining cells and split for each method and time point. Label untreated/treated cells for 4sU-seq with 4sU as described. Time points and samples for each method depend on the specific biological question being examined. Take samples for each time point and method, and follow the dedicated part of the protocol. Please click here to view a larger version of this figure.
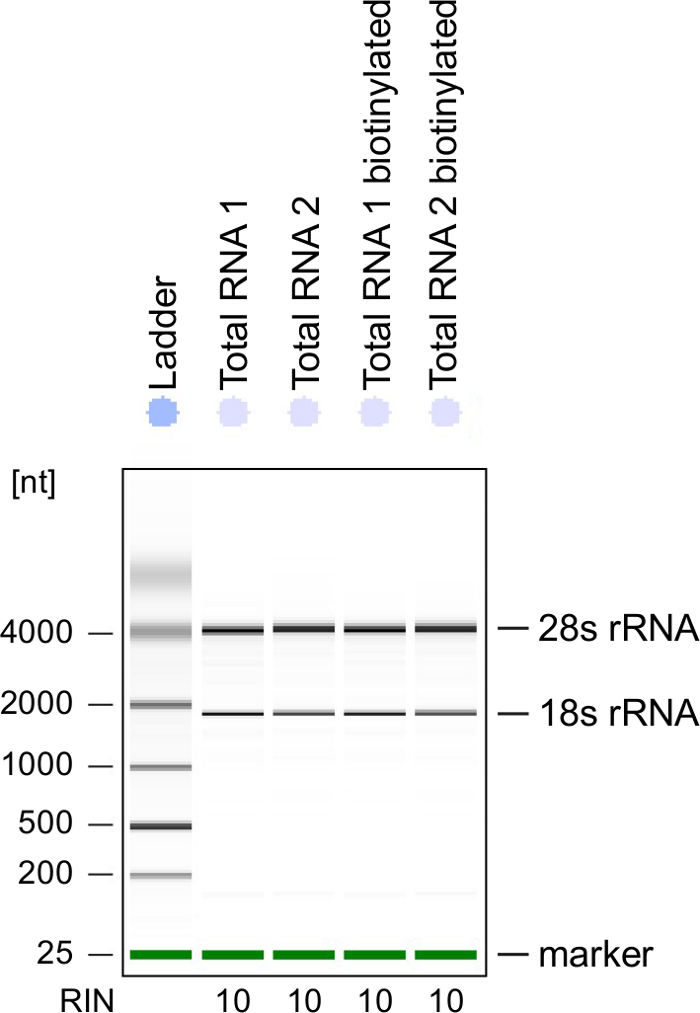
Figure 3: Quality control of 4sU-labeled RNA
Total RNA and biotinylated RNA obtained from activated Th1 cells were analyzed on a Bioanalyzer. 18S rRNA and 28S rRNA are shown and RNA integrity number (RIN) is given by the instrument to determine integrity of the RNA. RIN should be ≥8 to ensure RNA integrity. Please click here to view a larger version of this figure.
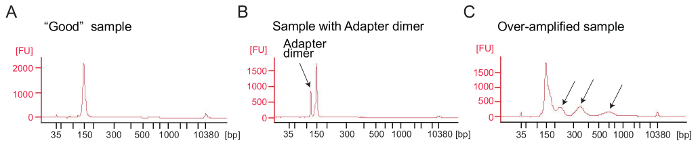
Figure 4: Bioanalyzer profiles of ribosome profiling libraries
(A) A good library: The sample shows a peak at the expected size range (140 – 160 bp) and no further purification is needed. (B) This sample shows excessive adapter dimer amplified product (120 bp) relative to the desired product (140 – 160 bp). This library requires further purification. (C) An over-amplified sample: Higher-than-expected molecular weight peaks and smeared PCR amplicons are visible (indicated by arrows). Please click here to view a larger version of this figure.
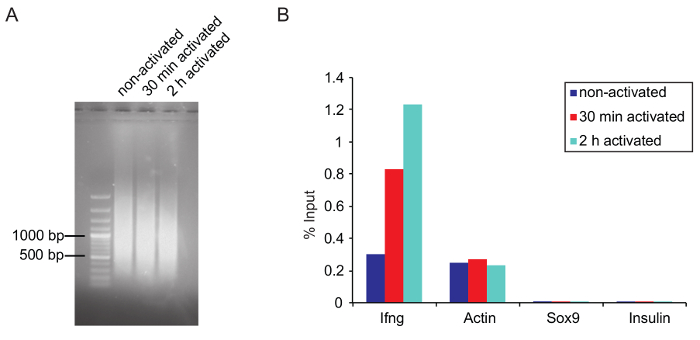
Figure 5: Optimal chromatin size after shearing and verification of ChIP by qPCR
(A) Agarose gel picture shows the optimal fragment size of the sheared chromatin from three samples that were sheared for 25 cycles on a sonicator and purified as described before in the protocol. (B) Q-PCR results of a total RNAPII ChIP (anti-RNA Pol II, 8WG16, ab817) is represented as a percentage of input. Ifng and actin primers were used as a positive while Sox9 and Insulin are negative controls (both genes are not expressed in activated Th1 cells). Please click here to view a larger version of this figure.
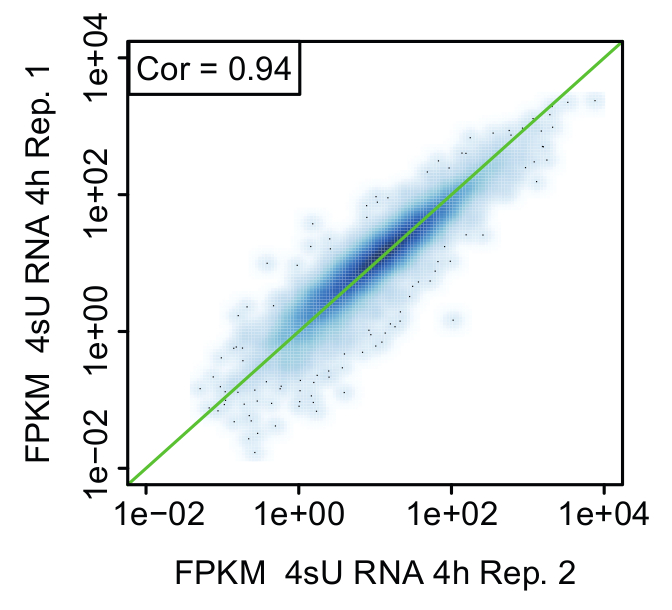
Figure 6: Comparison of biological replicates (Figure from Davari et al. 11)
Representative scatter plot comparing expression values (FPKM) between replicates of newly transcribed (4sU) RNA 4 h after stimulation of activated Th1 cells. The green line indicates equal FPKM values and rank correlation is indicated in each plot.
| Duration of labeling (min) | Recommended 4sU concentration (µM) |
| 120 | 100 – 200 |
| 60 | 200 – 500 |
| 15 – 30 | 500 – 1000 |
| <10 | 500 – 2000 |
Table 1: Recommended 4sU concentrations (from Rädle, et al.19)
The range of recommended 4sU concentrations is indicated for different times of labeling.
| Method | Cell number | RNA amount |
| 4sU-labeling | ≥2 x 107 | ≥60µg |
| Ribosome profiling | ≥2 x107 | |
| ChIP-seq | ≥2 x 107 - 3 x 107 |
Table 2: Required amount of primary T cells
Minimum amount of required primary T cells for each method. Amounts can be less when using other cell types.
Discussion
Analyzing the whole process of gene regulation is necessary to fully understand cellular adaptations in response to a specific stimulus or treatment. Combining total RNA-seq, 4sU-seq, ribosome profiling, and ChIP-seq at different time points leads to a comprehensive analysis of the main processes of gene regulation over time. A profound understanding of biological processes is required to define the experimental setup as well as optimal time points.
Since methods to study gene regulation improve rapidly, these protocols can be adapted to rapid changes. Nevertheless, they provide the most important methods for studying basic gene regulatory mechanisms in any type of cell. Here, we discuss some of the pitfalls and facts one has to consider when using these methods.
Cells: Cells must be highly viable and, if using primary isolated cells, purity of cell populations must be guaranteed (e.g., FACS analysis for primary T cells). Even slightly stressed cells can influence the results of these very sensitive sequencing methods and lower the amount of newly transcribed or translated RNA, and lead to unwanted readouts of the stress response in the sequencing results. The centrifugation speed mentioned in this protocol to pellet cells is optimized for primary T cells. Thus, adjust the speed according to the cell type.
4sU effects on cell physiology: In addition to the aforementioned options for verifying minimal perturbation to cellular physiology upon 4sU addition, further and/or additional analysis can be performed, especially when cell numbers are limited. Effects on cell proliferation can be tested by verifying the doubling time of cells by simply counting labeled and unlabeled cells. Nucleolar stress induction could also be tested by analyzing cell morphology via immunofluorescence staining of nucleolin and nuclei. To further verify the impact of 4sU, altered global gene expression could be measured by correlating read counts from labeled total RNA to unlabeled total RNA.
Cell numbers: For in vitro generated T cells, we recommend starting with at least the amount of cells indicated in Table 2. Choose adequate numbers per method according to the type of cells. Since T cells have less cytoplasm and RNA compared to other cells, most likely lower amounts of other cells will be adequate. For ChIP-seq, cell numbers highly depend on the antibody used and the expression level of the protein of interest within the cells. For histone or RNAPII ChIP, fewe cells can be used, whereas cell numbers need to be increased when transcription factors are used, especially if they are expressed at low levels.
4sU-labeling and RNA biotinylation: When using adherent cells, 4sU labeling can be directed as described by Rädle et al.19 Since cells very rapidly incorporate 4sU, it can be added directly to the medium of suspension, adherent, or semi-adherent cells.
It is recommended to start the biotinylation with 60 – 80 µg of RNA. Nevertheless, lower amounts of RNA can be used, although we did not test for less than 30 µg. Add a coprecipitant (e.g., GlycoBlue) when precipitating RNA if the pellet is hard to see. Duffy et al. have also shown that methylthiosulfonate-activated biotin (MTS-biotin) more efficiently reacts with 4sU-labeled RNA than HPDP-biotin31. Hence, it might be worth considering switching to MTS-biotin, in particular, for the recovery of small RNAs, which tend to have fewer uridine residues (refer to the biotinylation protocol mentioned by Duffy et al.; see Purification of 4sU-labeled RNA, Experimental Procedures).
For the recovery of newly transcribed RNA, it is possible to use paramagnetic beads or RNA Cleanup beads of your choice. Always take into account that these kits may or may not purify for specific RNAs. For example, if you are interested in miRNAs, consider using specific kits for miRNA capture and sequencing.
Quantification of newly transcribed RNA: To accurately quantify newly transcribed RNA, measurement should be performed by a suitable fluorometer (see Table of Materials). Within 1 h of 4sU exposure, newly transcribed RNA represents about 1 – 4% of total RNA. Newly transcribed RNA of 1 h labeled, activated T cells consists of ~90 – 94% of rRNA11.
Ribosome Profiling: When establishing the method, we determined that using 1.5x the amount of nuclease than suggested in the original protocol guarantees proper digestion. Also, no adverse effects have been reported for elevated amounts of nuclease. Since it is quite difficult to overdigest the RPFs while they are part of the RNA bound by ribosomal proteins, you can still slightly increase the amount to titrate optimal nuclease digestion.
If less than 500 ng of RPF RNA were recovered in step 4.4.2, repeat rRNA depletion and pool purified RPFs with RNA Clean & Concentrator-5 columns. Alternatively, load two identical samples next to each other on the gel (step 4.5.3) and pool gel slices during RNA elution from the gel (step 4.5.6).
We recommend cutting the RPFs on a gel as tightly as possible to the 28 and 30 nt bands. This helps in eliminating unwanted fragments from rRNAs and tRNAs, which later will become part of your library and reduce sequencing reads for your RPFs.
It is also recommended to avoid UV light during gel purification. This can create nicks in the RNA fragments, as well as pyrimidine dimers, that in the end can severely affect the library preparation and sequencing results.
Library preparation and sequencing of data: Ribosome profiling protocol enables to generate a cDNA library suitable for sequencing. Samples generated by 4sU-labeling can be directly used for library preparation with any appropriate RNA sequencing kit. Since newly transcribed RNA, especially when using short labeling times, may not yet be polyadenylated, no poly-A selection should be performed. Instead, we strongly recommend rRNA depletion to prevent reducing sequencing depth for the actual sample. Using T cells, we started with 400 ng of newly transcribed and total RNA (depending on the kit, see materials), performed rRNA depletion and reduced cycles for PCR amplification to minimize PCR bias. Library preparation can be performed with less starting material. To account for library complexity numbers of PCR cycles should be optimized.
For ChIP-seq there are also many kits available for library preparation. In our hands library preparation worked well starting with 2 ng of ChIP DNA (see materials for a suggestion on which kit to use). Be sure to check the indices for color balance during sequencing. We recommend a sequencing depth of ≥40 x 106 reads each for 4sU-seq, total RNA-seq, and ChIP-seq samples, and ≥80 x 106 reads for ribosome profiling samples. The sequencing depth depends on the sample and the downstream bioinformatics analysis and should be considered carefully. To analyze intronic reads for cotranscriptional splicing, 100 bp paired-end sequencing needs to be chosen.
Sequencing bias: Sequencing has become the gold standard when determining global changes in transcription, translation, or transcription factor binding. Within recent years, existing methods were pushed to their limits or new techniques were developed for sequencing increasingly small starting amounts of RNA. This requires amplification of cDNA, which introduces noise or bias. Recently, unique molecular identifiers (UMIs) were developed to experimentally identify duplicates introduced by PCR. Recently, it was shown that UMIs just mildly improve sequencing power and false discovery rate for differential gene expression32. Nevertheless, consider using unique molecular identifiers (UMIs) for all sequencing libraries to control for library complexity, especially when starting with low amounts of RNA and when many PCR cycles are needed.
Buffer and stock solutions: All buffers for 4sU-seq and ribosome profiling must be prepared under strict RNase-free conditions using nuclease-free water. It is recommended to buy pre-made nuclease-free NaCl, Tris-HCl, EDTA, sodium citrate, and water. To ensure nuclease-free conditions, an RNase decontaminating solution can be used to clean pipettes or surfaces. All buffers for ChIP-seq need to be at least DNase-free and can be stored at room temperature. Always add protease inhibitors and, optionally, phosphatase inhibitors right before use and keep on ice.
Bioinformatics: Analysis of all sequencing data (i.e., ChIP-seq, RNA-seq, and ribsosome profiling) involves quality control (e.g., using FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), adapter trimming (e.g., with cutadapt20) followed by mapping to the reference genome for the cells under study. For RNA-seq data (both total and 4sU-seq), as well as ribosome profiling data, a spliced RNA-seq mapper is required, such as ContextMap 221. For ChIP-seq data unspliced alignments, using BWA-MEM22 is sufficient. Gene expression can be calculated using the RPKM model (Reads per Kilobase of Exon per Million Fragments Mapped)1, after determining read counts per gene using a program, e.g. featureCounts23. For peak calling from ChIP-seq data, a number of programs are available, e.g., MACS24 or GEM25. Further downstream analyses can be performed in R26, in particular using tools provided by the Bioconductor project27.
Here, a major challenge in integrating 4sU- and total RNA levels and translational activity from ribosome profiling is normalization. A classical approach to address this problem is to normalize to levels of house-keeping genes. To reduce noise due to random fluctuations for individual house-keeping genes, it is recommended to not just use a few house-keeping genes but median levels for a larger set, e.g. the >3,000 house-keeping genes compiled by Eisenberg and Levanon28. For calculation of RNA turnover rates from ratios of 4sU- to total RNA, normalization is based on median turnover rates (e.g., assuming an RNA half-life of 5h)29. However, since this assumes no overall changes for house-keeping genes, we recommend using analysis approaches independent of normalization, e.g., correlation-based clustering of a time-series of the different data types to identify groups of genes with distinct behavior in transcription and translation during activation. For a detailed description on bioinformatical integration of the different data types, we refer to the original publication11.
Analysis of turnover rates and data integration: A recently published paper33 comparing half-lives determined by a multiplexed gene control (MGC) to global methods could show that half-lives correlated best with those obtained by metabolic labeling methods compared to other methods (e.g., general inhibition of transcription by drugs). However, it should be mentioned that differences between half-life calculations can arise and have been described 15,34. We account for most of the problems and differences that are introduced by the stress response due to prolonged 4sU exposure. Therefore, it is indispensable to exclude the stress response introduced by 4sU-labeling. To further validate turnover rates, we recommend the use of MGCs.
Additionally, a data set as generated here could also be used for a more integrative data analysis (e.g., regulation of long non-coding RNAs)35,36.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We thank Lars Dölken for advice to establish 4sU labeling for primary T cells; Elisabeth Graf and Thomas Schwarzmayr for critical help in library generations and sequencing; Dirk Eick and Andrew Flatley for providing RNAPII and T cell antibodies; N. Henriette Uhlenhaut and Franziska Greulich for help in library preparation for ChIP-seq; Caroline C. Friedel was supported by grants FR2938/7-1 and CRC 1123 (Z2) from the Deutsche Forschungsgemeinschaft (DFG); Elke Glasmacher was supported by the grant GL 870/1-1 from the Deutsche Forschungsgemeinschaft (DFG) and the German Center for Diabetes Research (DZD), Helmholtz Zentrum München.
Materials
| 4sU-labeling | |||
| 4-thiouridine (100 mg) | Carbosynth | 13957-31-8 | Prepare 50 mM stock in sterile H2O/PBS; store at –20°C in aliquots of 50-500 µl; do not refreeze. |
| 1.5 ml safe-lock tubes | Eppendorf | 30121589 | Optional |
| 1.5 ml screw-top polypropylene tubes | Sarstedt | 72692005 | Compatible with Dimethylformamide |
| 15 ml tubes | BD Falcon | 352096 | Compatible with Dimethylformamide |
| 2.0 ml screw-top polypropylene tubes | Sarstedt | 72694005 | Compatible with Dimethylformamide |
| 50 ml tubes | BD Falcon | 352070 | Compatible with Dimethylformamide |
| Agencourt RNAClean XP | Beckman Coulter | A63987 | We recommend to use these paramagnetic beads. Aliquot and store at 4°C |
| Chloroform | Sigma Aldirch | 372978 | WARNING – HAZARDOUS TO HEALTH |
| Dimethylformamide | Sigma Aldrich | D4551 | |
| Dithiothreitol (DTT) | Roth | 6908.1 | Prepare as 100 mM DTT in nuclease-free H2O; always prepare fresh |
| Ethanol | Merck | 1.00983.1000 | |
| EZ-Link Biotin-HPDP (50 mg) | Pierce | 21341 | Prepare 1 mg/ml stock solution by dissolving 50 mg Biotin-HPDP in 50 ml DMF. Gentle warming enhances solubilisation. Store at 4°C in aliquots of 1 ml. DMF dissolves some plastic materials. We recommend to use glass pipettes to transfer DMF from ist stock glass bottle to 50 ml Falcon tubes. |
| High Sensitivity DNA Kit | Agilent Technologies | 5067-4626 | |
| Isopropanol | Merck | 1.09634.1011 | |
| NaCl (5M) | Sigma Aldrich | 71386 | Stock solution |
| nuclease-free EDTA (500 mM ), pH 8.0 | Invitrogen | 15575-020 | Stock solution |
| Nuclease-free H2O | Sigma Aldrich | W4502 | Stock solution |
| nuclease-free Tris Cl (1M), pH 7.4 | Lonza | 51237 | Stock solution |
| Phase Lock Gel Heavy tubes (2.0 ml) | 5Prime | 2302830 | Use in step 1.3.4. |
| Polypropylene 15 ml centrifuge tubes | Greiner Bio-One | 188271 | Or equivalent; they have to tolerate up to 15,000 × g |
| QIAzol Lysis Reagent (200 ml) | Qiagen | 79306 | Use this or equivalent TRI reagent for RNA isolation, WARNING – CORROSIVE and HAZARDOUS TO HEALTH! Ensure immediate access to Phenol antidote (PEG-Methanol) |
| Qubit RNA HS assay kit | Life Technologies | Q32852 | Use this kit for quantifying RNA quantity in step 1.4.11 |
| RNeasy MinElute Kit | Qiagen | 74204 | Optional; includes Buffer RLT |
| Sodium citrat | Sigma Aldrich | C8532 | Prepare 1.6 M stock solution using nuclease-free water |
| Tween 20 | Sigma Aldrich | P1379 | |
| µMacs Streptavidin Kit | Miltenyi | 130-074-101 | Store at 4°C, includes µMacs columns used in step 1.4.6. (store at RT) |
| Cell viability and stress assay | |||
| PE Annexin V Apoptosis Detection Kit I | BD Biosciences | 559763 | Optional |
| Thapsigargin | Sigma-Aldrich | T9033 | Optional |
| p53 | abcam | ab26 | Optional |
| p-EIF2a (Ser51) | Cell Signaling | 9721 | Optional |
| BH3I-1 | Sigma-Aldrich | B 8809 | Optional |
| Buffers | |||
| 4sU Washing Buffer | store at RT | 100 mM Tris pH 7.4, 10 mM EDTA, 1 M NaCl, 0.1% Tween 20 in nuclease-free H2O | |
| Biotinylation Buffer (10x) | store at 4 °C | 100 mM Tris pH 7.4, 10 mM EDTA in nuclease-free water; make aliquots of 1 ml; store at 4°C | |
| RNA precipitation buffer | store at RT | 1.2 M NaCl, 0.8 M sodium citrate in nuclease free water. Prepare in advance under nuclease-free conditions. Store at room temperature in 50 ml falcon tubes. | |
| Equipment | |||
| 2100 Bioanalyzer instrument | Agilent | G2939BA | |
| RNA 6000 Nano Kit | Agilent | 5067-1511 | Use this kit to verify RNA integrity in step 1.3.10 |
| RNA 6000 Pico Kit | Agilent | 5067-1513 | Optional |
| UV/VIS spectrophotometer | Thermo Scientific | NanoDrop 1000 | Or equivalent. Use in step 1.2.2/3.1.8/3.3.3/3.4.3 |
| High-speed centrifuge | Thermo Scientific | Heraeus Multifuge X3R | Or equivalent equipment capable of reaching 13,000 × g |
| High-speed rotor | Thermo Scientific | Fiberlite F15-6 x 100y | |
| Adaptors for 15 ml tubes | |||
| Refrigerated table-top centrifuge | Eppendorf | 5430 R | Or equivalent. |
| Thermomixer | Eppendorf | Thermomixer C | Or equivalent. |
| Magnetic stand | Miltenyi Biotec | 130-042-109 | One stand holds 8 µMacs columns. |
| Ultra-fine scale | Mettler Toledo | ML204T | Or equivalent. |
| E-Gel iBase Power System | Invitrogen | G6400UK | For RNA gels; or equivalent. |
| E-Gel EX 1% agarose precast gels | Invitrogen | G4020-01 | For RNA gels; or equivalent. |
| DynaMag-2 Magnet-1 each | Life Technologies | 12321D | |
| RNaseZap | Sigma | R2020 | Optional |
| TruSeq stranded total RNA library prep kit | Illumina | RS-122-2201 | Or equivalent. For T cells we used 400 ng 4sU and Total RNA with 11 cycles for PCR amplification. rRNA depletion is included in this kit |
| Nanodrop | Thermo Scientific | use a Nanodrop or equivalent instrument to measure RNA concentration | |
| Ribosome Profiling | |||
| TruSeq Ribo Profile kit (Mammalian or Yeast) | Illumina | RPYSC12116 (Yeast) | |
| TruSeq Ribo Profile kit (Mammalian or Yeast) | Illumina | RPHMR12126 (Mammalian) | |
| Illustra MicroSpin S-400 HR Columns | GE Healthcare | 27-5140-01 | |
| RNA Clean & Concentrator-25 kit | Zymo Research | R1017 | |
| RNA Clean & Concentrator-5 kit | Zymo Research | R1015 | |
| Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) | Illumina | MRZG126 or MRZG12324 | |
| (High Sensitivity DNA Kit) | Agilent Technologies | 5067-4626 | Already needed for 4sU-seq |
| All other consumables and equipment are listed in the User guide | !!! | Carefully read the user guide and order required consumables in advance (consider a long delivery time for some consumables e.g. gels) | |
| ChIP | |||
| 10 mM Tris-HCl (pH 8.0) | gereral lab supplier | ||
| 100 bp Plus Marker | Thermo Fisher | SM0323 | |
| 16 % Formaldehyde | Thermo Fisher | 28908 | Add to a final concentration of 1 % |
| 70% EtOH | gereral lab supplier | Always prepare fresh | |
| Agarose | gereral lab supplier | ||
| Agencourt RNAClean XP beads | Beckman Coulter | A63987 | We recommend to use these paramagnetic beads. Aliquot and store at 4°C |
| ChIP library preparation kit | KapaBiosystems | KK8504 | Or use the kit of your choice |
| DNA low bind microcentrifuge tubes | Eppendorf | Z666548-250EA | or equivalent |
| Dynabeads Protein G | Invitrogen | 10004D | Use these superparamagnetic beads coupled to protein G in step 4.3.1.; Bring to RT before use |
| Glycine | gereral lab supplier | Prepare a 2M stock solution | |
| Glycogen | Roche | 10-901-393-001 | |
| MinElute PCR Purification Kit | Qiagen | 28004 | Use this kit (or equivalent) to purify chromatin in step 4.2.4. |
| Phosphatase Inhibitor (PhosStop) | Roche | 4906837001 | Add freshly to the buffer and keep on ice |
| Power SYBRgreen Master mix | Thermo Fisher | 4367659 | |
| Protease Inhibitor (cOmplete, EDTA-free) | Roche | 11873580001 | Add freshly to the buffer and keep on ice |
| Proteinase K | Invitrogen | 25530049 | |
| Qubit dsDNA HS Assay kit | Invitrogen | Q32851 | Use this kit for quantifying DNA quantity in step 4.6.4. on a Qubit Fluorometer |
| Rnase, DNase free | Roche | 11-119915001 | |
| Salmon sperm (sonicated to around 100bp) | Sigma | D1626 | |
| TE pH 8.0 | gereral lab supplier | ||
| Antibodies (ChIP grade if possible) | |||
| anti-RNA Pol II [8WG16] | abcam | ab817 | |
| anti-Histon H3K36me3 | abcam | ab9050 | |
| or antibody of interest | |||
| Buffers | |||
| Binding/Blocking buffer | Store at RT | PBS with 0.5 % BSA and 0.5 % Tween 20 | |
| Cell-Lysis buffer | Store at RT | 5 mM Pipes [pH 8.0], 85 mM KCl, and 0.5 % NP40 | |
| ChIP IP buffer | Store at RT | 0.01 % SDS; 1. 1% Triton X-100;1.2 mM EDTA; 16.7 mM Tris-HCl, pH 8.1; 16.7 mM NaCl | |
| Elution buffer | Store at RT up to 6 months | 10 mM Tris-HCl (pH 8.0), 5 mM EDTA (pH 8.0), 300 mM NaCl and 0.5 % SDS | |
| Nuclei-Lysis buffer | Store at RT | 50 mM Tris [pH 8.0], 10 mM EDTA, and 1 % SDS | |
| Wash buffer I | Store at RT | 0.1 % SDS; 1 % Triton X-100; 2 mM EDTA; 20mM Tris-HCL pH 8.1; 150 mM NaCl | |
| Wash buffer II | Store at RT | 0.1 % SDS; 1 % Triton X-100; 2 mM EDTA; 20 mM Tris-HCL pH 8.1; 500 mM NaCl | |
| Wash buffer III | Store at RT | 0.25 M LiCl; 1% NP-40; 1 mM EDTA; 10 mM Tris-HCl, pH 8.1 | |
| Equipment | |||
| 2100 Bioanalyzer instrument | Agilent | G2939BA | use this instrument for electrophoretical analysis |
| Nanodrop | Thermo Scientific | ||
| Bioruptor TBX microtubes 1.5 ml | Diagenode | C30010010 | |
| or tubes special for your sonication device | |||
| Bioruptor sonication device or sonication device of your choice | Sonication of T cells with Bioruptor: 20 – 25 cycles (30 s on, 30 s off at high in two 1.5 ml bioruptor microtubes with 500 µl each tube) | ||
| Magnetic stand for tubes | |||
| Thermomixer | |||
| Agarose gel electrophoresis | |||
| Qubit Fluorometer | Thermo Scientific | Use this Fluorometer for quantifying low amounts of RNA/DNA |
Referenzen
- Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 5 (7), 621-628 (2008).
- Chavez, S., Garcia-Martinez, J., Delgado-Ramos, L., Perez-Ortin, J. E. The importance of controlling mRNA turnover during cell proliferation. Curr Genet. 62 (4), 701-710 (2016).
- Rutkowski, A. J., et al. Widespread disruption of host transcription termination in HSV-1 infection. Nat Commun. 6, 7126 (2015).
- Ozsolak, F., Milos, P. M. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 12 (2), 87-98 (2011).
- Carninci, P., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 38 (6), 626-635 (2006).
- Churchman, L. S., Weissman, J. S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 469 (7330), 368-373 (2011).
- Core, L. J., Waterfall, J. J., Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 322 (5909), 1845-1848 (2008).
- Hah, N., et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 145 (4), 622-634 (2011).
- Dolken, L., et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 14 (9), 1959-1972 (2008).
- Paulsen, M. T., et al. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods. 67 (1), 45-54 (2014).
- Davari, K., et al. Rapid Genome-wide Recruitment of RNA Polymerase II Drives Transcription, Splicing, and Translation Events during T Cell Responses. Cell Rep. 19 (3), 643-654 (2017).
- Windhager, L., et al. Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res. 22 (10), 2031-2042 (2012).
- Park, P. J. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 10 (10), 669-680 (2009).
- Blecher-Gonen, R., Barnett-Itzhaki, Z., Jaitin, D., Amann-Zalcenstein, D., Lara-Astiaso, D., Amit, I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc. 8 (3), 539-554 (2013).
- Schwanhausser, B., et al. Global quantification of mammalian gene expression control. Nature. 473 (7347), 337-342 (2011).
- Larsson, O., Tian, B., Sonenberg, N. Toward a genome-wide landscape of translational control. Cold Spring Harb Perspect Biol. 5 (1), a012302 (2013).
- Brar, G. A., Weissman, J. S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol. 16 (11), 651-664 (2015).
- Hafner, M., et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 141 (1), 129-141 (2010).
- Radle, B., Rutkowski, A. J., Ruzsics, Z., Friedel, C. C., Koszinowski, U. H., Dolken, L. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J Vis Exp. (78), (2013).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 17 (1), 10-12 (2011).
- Bonfert, T., Kirner, E., Csaba, G., Zimmer, R., Friedel, C. C. ContextMap 2: fast and accurate context-based RNA-seq mapping. BMC Bioinformatics. 16, 122 (2015).
- Li, H., Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26 (5), 589-595 (2010).
- Liao, Y., Smyth, G. K., Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30 (7), 923-930 (2014).
- Zhang, Y., et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9 (9), R137 (2008).
- Guo, Y., Mahony, S., Gifford, D. K. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 8 (8), e1002638 (2012).
- Team, R. D. C. . R: A Language and Environment for Statistical Computing. , (2016).
- Huber, W., et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods. 12 (2), 115-121 (2015).
- Eisenberg, E., Levanon, E. Y. Human housekeeping genes, revisited. Trends Genet. 29 (10), 569-574 (2013).
- Friedel, C. C., Dolken, L. Metabolic tagging and purification of nascent RNA: implications for transcriptomics. Mol Biosyst. 5 (11), 1271-1278 (2009).
- Burger, K., et al. 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 10 (10), 1623-1630 (2013).
- Duffy, E. E., Rutenberg-Schoenberg, M., Stark, C. D., Kitchen, R. R., Gerstein, M. B., Simon, M. D. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol Cell. 59 (5), 858-866 (2015).
- Parekh, S., Ziegenhain, C., Vieth, B., Enard, W., Hellmann, I. The impact of amplification on differential expression analyses by RNA-seq. Sci Rep. 6, 25533 (2016).
- Baudrimont, A., et al. Multiplexed gene control reveals rapid mRNA turnover. Sci Adv. 3 (7), e1700006 (2017).
- Rabani, M., et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 29 (5), 436-442 (2011).
- Schlackow, M., Nojima, T., Gomes, T., Dhir, A., Carmo-Fonseca, M., Proudfoot, N. J. Distinctive Patterns of Transcription and RNA Processing for Human lincRNAs. Mol Cell. 65 (1), 25-38 (2017).
- Mukherjee, N., Calviello, L., Hirsekorn, A., de Pretis, S., Pelizzola, M., Ohler, U. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat Struct Mol Biol. 24 (1), 86-96 (2017).

