Automatically Generated
Longitudinal In Vivo Imaging of Retinal Cells with Tailored Adeno-Associated Virus Serotypes via Confocal Scanning Laser Ophthalmoscopy
Summary
This protocol describes the use of adeno-associated virus (AAV) vectors for cell-specific labeling and in vivo imaging using a confocal scanning laser ophthalmoscope (CSLO). This method enables the investigation of different retinal cell types and their contributions to retinal function and disease.
Abstract
The dynamic nature of retinal cellular processes necessitates advancements in gene delivery and live monitoring techniques to enhance the understanding and treatment of ocular diseases. This study introduces an optimized adeno-associated virus (AAV) approach, utilizing specific serotypes and promoters to achieve optimal transfection efficiency in targeted retinal cells, including retinal ganglion cells (RGCs) and Müller glia. Leveraging the precision of confocal scanning laser ophthalmoscopy (CSLO), this work presents a non-invasive method for in vivo imaging that captures the longitudinal expression of AAV-mediated green fluorescent protein (GFP). This approach eliminates the need for terminal procedures, preserving the continuity of observation and the well-being of the subject. Furthermore, the GFP signal can be traced in AAV-infected RGCs along the visual pathway to the superior colliculus (SC) and lateral geniculate nucleus (LGN), enabling the potential for direct visual pathway mapping. These findings provide a detailed protocol and demonstrate the application of this powerful tool for real-time studies of retinal cell behavior, disease pathogenesis, and the efficacy of gene therapy interventions, offering valuable insights into the living retina and its connections.
Introduction
Being the only optically accessible part of the central nervous system, the retina serves as a valuable model for neuroscience research1. Retinal ganglion cells (RGCs), the output neurons of the retina that transmit visual information to the brain, play a crucial role in visual function. Their loss or dysfunction leads to vision impairment and irreversible blindness, as seen in glaucoma and other optic neuropathies2. Müller glia, the principal glial cells in the retina, are essential for maintaining retinal homeostasis, providing structural and metabolic support to neurons, regulating neurotransmitter levels, and contributing to retinal repair and regeneration3. Their dysfunction is implicated in various retinal diseases, including diabetic retinopathy4, age-related macular degeneration5, and ocular ischemic syndrome6. RGCs and Müller glia exhibit close interactions and interdependence; Müller glia provide essential support to RGCs, while RGC activity can influence Müller glia function3,7. Studying both RGCs and Müller glia is crucial for understanding retinal function and developing effective treatments for multiple retinal diseases.
Current assessments in retinal research primarily utilize techniques like optical coherence tomography (OCT) to measure the thickness of the retinal nerve fiber layer or the trajectories of axon bundles8,9. While these methods are invaluable for detecting RGC loss, they do not provide a detailed view of RGC morphology and glial cells due to limited resolution. Similarly, although advanced techniques like adaptive optics scanning laser ophthalmoscopy (AO-SLO) enable cellular-level imaging of RGCs, photoreceptors, and glial cells in the living human retina10, their technical complexity and limited accessibility confine their use primarily to specialized research settings. Given these constraints, there is an ongoing need for developing more accessible and reliable methods for the in-depth study of specific retinal cell populations in vivo.
Accordingly, this protocol aims to introduce an alternative imaging approach suited for research applications in retinal cells. It combines the power of AAV-mediated cell-type-specific labeling with the non-invasive nature of CSLO imaging. Adeno-associated viruses (AAVs) are versatile gene delivery vectors known for their low immunogenicity and ability to transduce a broad range of cell types, including both dividing and non-dividing cells11. This makes them ideal tools for targeting specific cell populations within the complex retinal environment. By utilizing AAV vectors with carefully selected serotypes and promoters, selective expression of fluorescent proteins can be achieved in multiple cell types of interest, such as RGCs and Müller glia. For example, AAV2 is known for its higher transduction efficiency in RGCs12,13, while AAV8 is markedly effective at targeting photoreceptors14, and AAV9 demonstrates strong transfection capabilities in Müller glia15, showing broad efficiency across various retinal cell layers. It is important to note that the effectiveness of AAV relies not only on the choice of serotype but also on the promoters, which dictate the intensity and cell specificity of transgene expression, underscoring the importance of careful selection to achieve optimal transduction.
For RGC labeling, this protocol employs AAV2 with the human synapsin (hSyn) promoter. AAV2 exhibits efficient transduction of RGCs following intravitreal injection13, and the hSyn promoter, a ubiquitous neuronal promoter, drives strong and specific transgene expression within these cells16. For Müller glia, the protocol utilizes AAV9 vectors driven by the GfaABC1D promoter17, which demonstrates strong transgene expression in these cells15. This targeted labeling approach enables researchers to distinguish these cells from the surrounding retinal tissue and track them over time, providing a basis for in vivo surveillance of retinal cells and their responses to bio-environmental changes.
Confocal scanning laser ophthalmoscopy (CSLO) is a non-invasive imaging technique that provides high-resolution images of the living retina, enabling real-time visualization of fluorescently labeled retinal cell populations18,19,20. A focused laser beam scans across the retina, capturing emitted light that passes through a pinhole to eliminate out-of-focus signals, resulting in sharper images with enhanced contrast. This protocol utilizes a Heidelberg Spectralis CSLO system, which has been widely used for retinal cell imaging in live animals, including studies visualizing transgenic-labeled RGCs21,22and microglia23. By employing the HRA CSLO unit with a 488 nm laser and appropriate filters, researchers can image fluorescently labeled RGCs or Müller glia in live animals following intravitreal injection of AAV vectors carrying fluorescent reporter genes. The longitudinal imaging protocol, with weekly sessions covering both central and peripheral retina, tracks changes over time. To prioritize animal welfare, the protocol utilizes the automatic eye-tracking system (ART) of the HRA CSLO unit, enabling precise image acquisition without the need for general anesthesia or contact lenses.
This protocol harnesses the combined power of AAV and CSLO to enable the longitudinal monitoring of specific retinal cell types in vivo. By pairing the cell-type specificity of AAV-mediated labeling with the non-invasive, high-resolution imaging capabilities of CSLO, this method allows researchers to study the dynamic changes in RGCs and Müller glia in response to various stimuli or interventions. These insights hold significant potential for informing the development of new diagnostic and therapeutic strategies for retinal diseases.
Protocol
All experiments were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the Institutional Animal Care and Use Committee of Capital Medical University, Beijing. Four-week-old adult male C57BL/6J mice (weighing between 15-20 g) were used for all experiments and housed in temperature-controlled rooms with a 12/12-h light/dark cycle. Standard rodent chow and water were available ad libitum. The details of the reagents and equipment used in this study are listed in the Table of Materials.
1. AAV-mediated retinal cell transfection
NOTE: For targeted transduction of RGCs, this protocol uses AAV2-hSyn-eGFP, which incorporates the hSyn promoter to drive robust expression of enhanced GFP. Müller glia is targeted using AAV9-GfaABC1D-eGFP. To achieve optimal transduction efficiency and robust transgene expression, a minimum AAV titer of 1 x 1012 viral genomes (vg)/mL is recommended for intravitreal injections in mice13.
- Adhere to stringent safety protocols when conducting AAV-mediated retinal cell transfection procedures to prevent accidental exposure and ensure a safe working environment.
- Perform all procedures involving viral vectors within a certified biosafety cabinet (BSC). Equip personnel with appropriate personal protective equipment (PPE), including lab coats, gloves, and eye protection, throughout the duration of the procedure.
- Dispose of all sharps and biohazard materials properly in accordance with institutional safety protocols to maintain compliance and safeguard all laboratory personnel.
2. Preparation of viral vectors and animals
- Obtain AAV2-hSyn-eGFP or the AAV9-GfaABC1D-eGFP vectors stored at -80 °C. Thaw vectors on ice immediately before use to preserve viral integrity.
- Anesthetize the mice with an intraperitoneal injection of pentobarbital sodium at a dosage of 50 mg/kg. Confirm the depth of anesthesia by testing the rear foot reflexes before further actions.
- Trim the whiskers to prevent interference with the surgical area during the procedure. Disinfect the orbit with a 20% Povidone-iodine solution for 2 s, then rinse thoroughly with sterile normal saline.
- Apply a drop of 0.5% proparacaine topically to minimize discomfort and prevent involuntary eye movements.
3. Handling and intravitreal injection of viral vectors
- Transfer 1 µL of selected AAV solution (either AAV2-hSyn-eGFP or AAV9-GfaABC1D-eGFP) to a piece of clean, prechilled paraffin film to avoid temperature fluctuations that might affect the viral activity.
- Aspirate the AAV solution using a Hamilton glass syringe with a 33 G beveled needle under the ophthalmic surgical microscope.
- Place the mouse in ventral recumbency under the microscope. Tilt the head slightly to elevate the ocular side intended for injection. Adjust and optimize the magnification of the microscope to clearly identify the superior region of the mouse orbit.
- Insert the needle 1-2 mm posterior to the limbus at the superior scleral margin of the eye into the vitreous cavity. Inject the AAV solutions slowly to avoid backflow or pressure-induced damage.
NOTE: Injection site location: Perform the injection in the superior temporal quadrant of the eye, approximately 1 mm posterior to the limbus. Choose this location to avoid major blood vessels and minimize the risk of lens damage. Avoid injection sites too close to the limbus to prevent iris penetration or lens abrasion. Avoiding vasculature: Use an operating microscope to carefully examine the planned injection site for visible blood vessels prior to injection. If vessels are present, slightly adjust the entry point to avoid them. Insert the needle at a shallow angle, parallel to the iris, to further reduce the risk of vascular damage. Avoid repeated injections in the same area to reduce the risk of subretinal hemorrhage. - Keep the needle in place for 30 s to allow the vectors to disperse within the vitreous chamber and settle onto the retinal surface, thereby maximizing the chances of successful transduction.
- Withdraw the needle slowly to minimize the risk of vitreous backflow.
- Apply a topical antibiotic ointment to the injection site immediately after injection. Ensure that the ointment is applied uniformly to fully cover the ocular surface, preventing ocular infection and inflammation.
NOTE: The antibiotic ointment contains 1 mg/g Dexamethasone, 3500 IU/g Neomycin Sulphate, and 6000 IU/g Polymyxin B Sulphate. - Place the mouse on a heating pad to maintain body temperature during recovery from anesthesia, and closely monitor them for any signs of distress or abnormalities. Once fully recovered and exhibiting no complications, return them to their cages with access to normal food and water.
4. In vivo imaging with CSLO
- Animal preparation
- Select the mouse infected with AAV2-hSyn-eGFP or AAV9-GfaABC1D-eGFP after 4 weeks of intravitreal injection. Apply one drop of an eye solution containing 0.5% tropicamide and 0.5% phenylephrine to the eye surface to dilate the pupil to approximately 2 mm in diameter22.
NOTE: A minimum of 4 weeks of AAV transduction time interval is recommended for the optimal retinal cells fluorescent labeling post intravitreal injection13. Additionally, in our optic nerve crush (ONC) experiments, the retinas were imaged at 4 weeks post-infection, as well as on day 7 and 14 following the crush injury. These time points were chosen to capture both the initial AAV expression and the progression of retinal changes after ONC. - Cover the imaging platform with a piece of the clean pad to avoid the possible contamination of animal excrement during the operation process.
NOTE: The custom-built imaging platform provides ample space for the animal to lie prone, ensuring stability and minimizing movement during manual restraint for optimal image acquisition. - Carefully transfer the mouse onto the imaging platform and allow an acclimatization period of 2-5 min prior to imaging to minimize stress and facilitate adaptation to the imaging environment.
- With the assistance of a second technician, gently restrain the animal while maintaining a conscious state throughout the procedure. Provide a 10 s rest interval after 15 s of imaging to permit eye blinking and maintain corneal moisture.
NOTE: Gently scrub the scalp skin to induce spontaneous eyelid lifting, avoiding the use of forced eyelid restraint or additional devices such as clips, thereby reducing the risk of eye injury. General anesthesia is not employed to preserve optical clarity during extended imaging sessions, as it has the potential to exacerbate transient lens opacity. Balance high-quality image acquisition with animal comfort and safety throughout the procedure. Complete the entire procedure within 5 min to ensure animal well-being and maintain imaging quality. - Adjust the head posture and align the camera lens by fine-tuning the focus knob (Figure 1B) and micromanipulator (Figure 1C) to aim the region of interest (ROI) for subsequent in vivo imaging.
- Select the mouse infected with AAV2-hSyn-eGFP or AAV9-GfaABC1D-eGFP after 4 weeks of intravitreal injection. Apply one drop of an eye solution containing 0.5% tropicamide and 0.5% phenylephrine to the eye surface to dilate the pupil to approximately 2 mm in diameter22.
- System configuration
- Attach a non-contact 55° wide angle lens on the camera to expand the field of view of the fundus area.
- Set the filter wheel to position A (angiography) to enable the acquisition of infrared (IR) and fluorescent angiography (FA) imaging mode (Figure 1A). Turn on the laser and power supply to activate the CSLO imaging system.
- Open the software Heidelberg Eye Explorer and create a new examination by clicking on the New Patient icon at the top of the menu bar. Enter the relevant animal information and accept the default corneal curvature of 7.7 mm in the pop-up window.
- Select the yellow Start button (Figure 1E) on the control panel screen to initiate the live imaging mode.
- In vivo CSLO imaging
- Select the IR mode (Figure 1G) with autofluorescence at 820 nm excitation wavelength on the control panel. To achieve high-resolution imaging, the High Res. (HR) mode is activated either through the window interface or the manual control panel (Figure 1F).
NOTE: Infrared reflectance (IR) imaging is typically the first step in CLSO ophthalmic imaging, acquired before fluorescein angiography (FA), to provide a baseline view. Even illumination, minimal artifacts, and sharp focus on major retinal vessels are crucial for the reliability of z-plane focusing and the achievement of high-quality IR images. - Move the lens towards the mouse eye using the joystick with the XYZ-micromanipulator for fine adjustment. Examine the optical transparency and turn the sensitivity knob (Figure 1D) counterclockwise to decrease the brightness of the fundus image to 40%-60%, thereby avoiding overexposure of the fundus and enhancing visualization of retinal details.
- Adjust the focus knob and micromanipulator until the optic disc and retinal vessels are unambiguously identified on the fundus.
NOTE: Criteria of the optimal focal plane: (1) Evenly illuminated fundus view without dark corners. (2) Uniformed illumination surrounding the optic disc without focal dark spots or distortion. (3) Clear lumen shape of the large retinal vessels with the visible movement of blood flow. - Switch to the FA mode (Figure 1H) with a solid-state blue laser at an excitation wavelength of 488 nm and a barrier filter at 500 nm. Turn the sensitivity knob (Figure 1D) clockwise to increase the brightness of the fundus image to 50%-107% to illuminate the targeted retinal area.
- Set the ART mean value (a blue bar at the bottom left of the software interface) to at least 15 to enhance the signal-to-noise ratio. This function averages a series of B-scans acquired at the same location, improving image quality.
NOTE: While a higher ART averaging value generally enhances image quality through averaging, it also extends scan duration. This extended duration could potentially increase the risk of corneal dehydration and animal fatigue, factors that need to be carefully considered, especially in awake animal imaging. - Re-align to the inner plane of the retina, specifically targeting the GFP expressing RGCs or the Müller glial cells. Fine adjustments are made to ensure sharp illumination of the desired cell population, such as RGC soma and axons, or the Müller cell body. Imaging sensitivity is balanced to achieve optimal visualization and prevent oversaturation of the GFP-positive cells.
NOTE: To obtain the images with comparable clarity and brightness of the GFP signal, the sensitivity acquisition should be constant for all mice in the experiment. - Press down the sensitivity knob (Figure 1D) to initiate the ART mode, which generates a live mean image online. Wait until the blue bar (representing the ART Mean value) at the bottom left of the software interface has reached the set ART value (≥ 20 frames) and tap the Acquire button on the control interface to capture the fundus images.
NOTE: For ART mode, eye-tracking is incorporated to automatically compensate for minor eye movements during in vivo imaging, enabling consistent focus on the selected retinal focal plane. - Once acquired the desired images, exit the ART mode by pressing the sensitivity knob on the touch panel.
NOTE: To prevent corneal drying and allow for spontaneous eye blinking, 10 s breaks were implemented every 15 s during the imaging process. - To navigate the nasal and temporal retinal areas, move the camera head horizontally while observing the live image on the screen. Once the target region is reached, fine-tune the focus and start the imaging process as described in steps 2.3.1-2.3.8.
NOTE: Maintain slow and controlled camera movements while ensuring consistent and bright illumination of the fundus. If dark areas appear, utilize the micromanipulator to adjust the camera position vertically and re-center the retina. - To image the superior retina, gently adjust the mouse head posture to a face-up position. Moderately tilt the camera head upwards to align with the superior retinal region. Re-adjust the focus as needed and proceed with imaging.
- Upon completion of imaging, all desired retinal regions, exit the acquisition window, and the images are automatically saved. Apply topical lubricant on the imaged eye to maintain corneal hydration.
- To Initiate the examination of the contralateral eye, reposition the animal to the opposite side of the platform and repeat steps 2.3.1-2.3.11.
- Select the IR mode (Figure 1G) with autofluorescence at 820 nm excitation wavelength on the control panel. To achieve high-resolution imaging, the High Res. (HR) mode is activated either through the window interface or the manual control panel (Figure 1F).
5. Image processing and analysis
- Image export and preparation
- Open the Heidelberg Eye Explorer software and select the desired CSLO imaging session with good quality. Choose single or consecutive images of the same retinal location with consistent sensitivity settings over time.
- Exclude blurry images with poor focus or corneal clarity, ensuring that only images with clear visualization of retinal structures are used for analysis. Export these images in TIFF format for further processing.
- Image alignment and cropping
- Group images per mouse eye based on the same retinal region.
NOTE: The original images acquired from the CSLO system are saved in the following format: TIFF, with a dimension of 1536 x 1536 pixels, a resolution of 5.69 um/ pixel, and an ART value of ≥15. - For follow-up examinations, import images into suitable image processing software. Rotate and align the images as necessary to match the orientation of the baseline image. When saving or exporting the processed images, use settings that maintain the highest possible image quality to minimize potential compression artifacts.
- Align the CSLO images from a time series of the same eye by rotating them to match the respective baseline image using the vasculature and optic disc as landmarks.
NOTE: Carefully examine each aligned image, and before proceeding to crop, ensure they meet the following criteria: (1) Full retinal region capture: The image must fully encompass the intended retinal region of interest, including key landmarks like the optic disc and major blood vessels; (2) Absence of motion artifacts: The image should be free from blurring, streaking, or duplication of structures caused by eye movement during imaging; (3) Minimal distortion: The image should accurately represent the retinal structure without warping, stretching, or compression artifacts that could arise from imaging angles or lens effects. (4) Balance of image quality metrics: Assessments of background noise levels (Standard deviation of pixel intensities in structure-free areas < 5 (0-255 scale)); signal-to-noise ratio (SNR >20), illumination consistency (Coefficient of variation among six image quadrants <10%) and image uniformity (Coefficient of variation in homogeneous regions <15%). - Select the rotated image and export it in TIFF format. For each time point, use the cropping tool to randomly select and crop 5-10 areas (300 x 300 pixels each) containing GFP-positive cells from the quality-approved images. Export each cropped image individually in TIFF format for further analysis.
- Group images per mouse eye based on the same retinal region.
- Fluorescence intensity measurement
- Open the cropped CSLO images (8-bit grayscale, 300 x 300 pixels) in ImageJ/Fiji.
- To examine overall changes in fluorescent signals, measure the total fluorescence intensity per cropped image using Analyze > Measure (or Ctrl + M). Record the "Mean" value, representing the average GFP fluorescence intensity of the whole cropped image.
- RGC quantification
- In ImageJ/Fiji, adjust the intensity threshold of the cropped CSLO images using Image > Adjust > Brightness/Contrast to achieve optimal visualization of individual white soma bodies against the black background.
NOTE: Determine an individualized threshold after reviewing the batch of cropped images, or apply a consistent thresholding algorithm (e.g., Huang's method5 in ImageJ/Fiji). For Huang's method: (1) Convert images to 8-bit grayscale (Image > Type > 8-bit). (2) Access the thresholding tool (Image > Adjust > Threshold). (3) Select Huang as the thresholding method. (4) Preview the segmentation and click on Apply. The threshold will be automatically calculated and displayed on the image histogram. - Activate the Cell Counter plugin and click on each GFP-positive RGC soma for counting. Record the total number of cells marked in the analyzed region. Repeat this process for all cropped images from each follow-up time point.
- In ImageJ/Fiji, adjust the intensity threshold of the cropped CSLO images using Image > Adjust > Brightness/Contrast to achieve optimal visualization of individual white soma bodies against the black background.
- Data analysis
- Export the cell count and fluorescence intensity data to a spreadsheet or statistical software (e.g., GraphPad Prism).
- Perform appropriate statistical tests based on the corresponding experimental design and hypotheses. Interpret the results and conclude.
Representative Results
Following the presented protocol, different retinal cells were successfully visualized and tracked in vivo using a combination of AAV-mediated gene delivery and CSLO. AAV2-hSyn-eGFP effectively transduced RGCs, resulting in robust eGFP expression throughout the retina, as confirmed by CSLO and colocalization with the RGC-specific marker, RNA binding protein with multiple splicing (RBPMS), specifically found in the ganglion cell layer (Figure 2 and Figure 3). The protocol also confirmed the ability to target Müller glia using AAV9-GfaABC1D-eGFP. CSLO visualization revealed widespread eGFP expression, specifically within the inner retinal layers, consistent with the known major distribution segment of Müller glia. The GFP positive signal also highly colocalized with the Müller glia marker glutamine synthetase (GS), further confirming the specificity of this approach (Figure 2E,F and Figure 3B). These findings reaffirmed the versatility of AAV-aided gene delivery for studying different retinal cell types. To confirm the ability to target Müller glia, AAV9-GfaABC1D-eGFP was used. CSLO imaging revealed widespread eGFP expression, specifically within the inner retinal layers, consistent with the known distribution of Müller glia (Figure 2E,F). This selective expression pattern was further validated by the high degree of colocalization observed between the GFP signal and the Müller glia marker glutamine synthetase (GS) (Figure 3B).
To quantify the specificity and efficiency of the viral targeting approach for both RGCs and Müller glia, a colocalization analysis was performed to compare GFP fluorescence with standard cell-type markers (Figure 3B,D). A total of 6 mouse eyes were analyzed. For RGCs targeted with AAV2-hSyn-GFP, transduction efficiency was 64% ± 3%, while transduction specificity was 68% ± 3%. For Müller glia targeted with AAV9-GfaABC1D-GFP, transduction efficiency was 58% ± 2%, and transduction specificity was 71% ± 2%. These results demonstrate that this AAV-mediated gene delivery approach enables selective labeling of distinct retinal cell types, highlighting its versatility for studying cellular and molecular processes in the retina.
To assess the suitability of CSLO for longitudinal monitoring of retinal cell perturbations, a transient ONC model was performed in AAV2-hSyn-eGFP-infected mice. A progressive decline in eGFP signal and RGC numbers was observed over two weeks (Figure 4). RGC survival was quantified with three metrics: CSLO fluorescence intensity, in vivo cell counts for CSLO images, and ex vivo RGC counts of RBPMS-positive cells. After image processing as described in step 4, at least 10 regions of interest (ROIs) were cropped from each retina (6 eyes total), ensuring each ROI encompassed the same retinal location and exhibited clear cell signals at each time point. CSLO fluorescence intensity was then quantified by measuring the mean intensity within each cropped ROI at each time point. In vivo, cell counts were obtained by manually counting individual GFP-positive RGCs within the same cropped ROI at each time point. Lastly, retinas were harvested for RBPMS immunostaining, and RBPMS-positive cells were counted within 24 defined areas imaged per retinal sector for each retina. Significant negative correlations were found between all three RGC survival metrics (CSLO fluorescence intensity, in vivo cell counts, and ex vivo RBPMS-positive cell counts) and the time post-ONC (p < 0.001 for all, Figure 4C), demonstrating progressive RGC loss and degeneration. Importantly, no significant differences were observed across the different experimental groups in terms of initial RGC density or rate of decline, indicating a gross consistency in the ONC model and imaging approach. These findings are consistent with previous studies on RGC damage after ONC and validate the effectiveness of this CSLO-based protocol as a non-invasive and reliable tool for monitoring RGC health.
To further investigate RGC axonal projections within the visual pathway, a dual-labeling approach was used with the combination of AAV-mediated eGFP labeling with the anterograde tracer cholera toxin subunit B (CTB)24. Mice received an intravitreal injection of CTB and were sacrificed 2 days later for histological analysis. Colocalization of eGFP and CTB signals in optic nerves confirmed the positive tracing of RGC axons (Figure 5A). To further study the changes of signals after axonal injury, mice were subjected to unilateral optic nerve crush (ONC), a model of acute RGC damage25. Briefly, mice were anesthetized, and one optic nerve was exposed and crushed approximately 1 mm behind the globe using fine forceps for 5 s, inducing significant RGC axotomy and subsequent degeneration. Following ONC injury, a marked reduction in both eGFP and CTB intensity was observed, indicating substantial nerve fiber loss and axonal transport disruption. Further analysis of brain sections revealed RGC projections to the Superior Colliculus (SC), a midbrain structure involved in sensorimotor integration and eye movements, and the Lateral Geniculate Nucleus (LGN), the primary relay center for visual information to the visual cortex. After ONC, a decreased GFP intensity was revealed in the contralateral side of the injury (Figure 5B–E). These findings demonstrate the utility of this method for studying RGC axonal degeneration and mapping their projection patterns in vivo, offering a potential alternative to traditional invasive tracing methods.
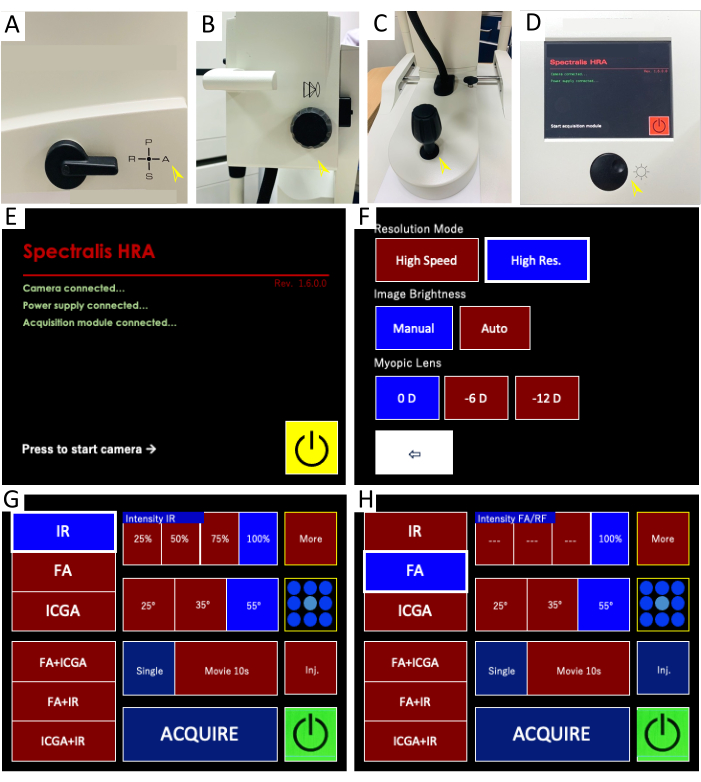
Figure 1: Confocal scanning laser ophthalmoscope (CSLO) imaging system and control panel interface for live retinal imaging. (A–D) Imaging system components: These images depict the key components of the HRA CSLO device. (A) The filter lever is set to position "A" for combined infrared reflectance (IR) and fluorescein angiography (FA) imaging. (B) The focus knob adjusts the fine focus of the objective lens. (C) The micromanipulator allows for precise positioning, alignment, and illumination of the light beam to the retina through multiple degrees of freedom (e.g., X, Y, Z axes) and rotation. (D) The sensitivity knob controls the brightness of both IR and FA images by adjusting the amplification of the detected light signal. Pressing the knob initiates image averaging, which combines multiple frames to reduce noise and enhance the signal-to-noise ratio. (E–H) Control panel interface: The screenshots display the CSLO control panel with the settings used for live image acquisition and visualization. (E) The acquisition control panel is initiated. (F) High-resolution mode is recommended for optimal image quality. (G) The IR imaging settings are highlighted, allowing visualization of retinal structures and blood vessels. (H) The FA imaging settings display options for both single-point and multi-point fluorescence imaging, enabling the detection of fluorescent signals within retinal cells. Please click here to view a larger version of this figure.
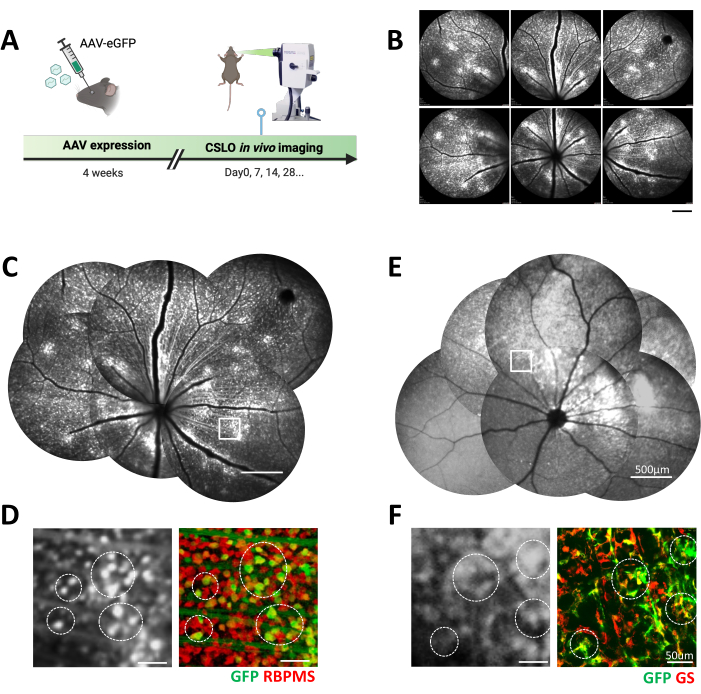
Figure 2: In vivo CSLO imaging of fluorescently labeled retinal cells following AAV transfection in C57 mice. (A) The experimental timeline of CSLO-aided longitudinal imaging after AAV transfection in the mouse eye. (B) Representative CSLO images of the superior and central regions of a C57 mouse retina after 4 weeks of AAV2-hSyn-eGFP expression. Scale bar: 500 µm. (C) A manually stitched retinal montage from (B) showcases the distribution of eGFP-expressing retinal cells. Scale bar: 500 µm. (D) A magnified CSLO image of the boxed area in (C) and the corresponding confocal microscopy image of a retinal flatmount. The colocalization of eGFP (green) with the RGC marker RBPMS (red) confirms AAV-mediated expression in retinal ganglion cells (examples circled). Scale bar: 50 µm. (E) A manually stitched montage of a C57 mouse retina 4 weeks post-injection with AAV9-GfaABC1D-eGFP, demonstrating Müller glia-specific eGFP expression. Scale bar: 500 µm. (F) A magnified CSLO image of the boxed area in (E) and the corresponding confocal microscopy image of a retinal flatmount. The colocalization of eGFP (green) with the Müller glia marker glutamine synthetase (GS, red) confirms AAV-mediated expression in Müller glial cells (examples circled). Scale bar: 50 µm. Please click here to view a larger version of this figure.
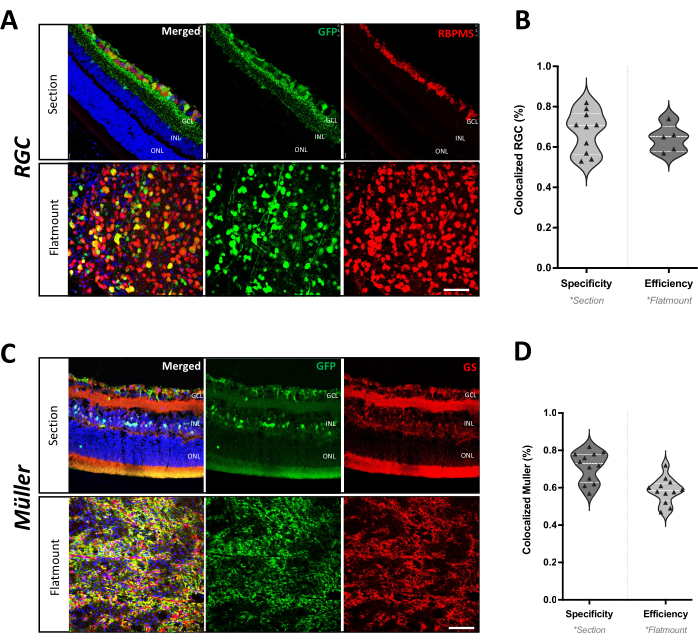
Figure 3: Cell type specificity of AAV-mediated gene delivery in mouse retina. (A) Immunohistochemistry of a retinal section and flatmount from C57 mice after 4 weeks of AAV2-hSyn-eGFP infection. The colocalization of GFP (green) with RBPMS (red) confirms RGC-specific expression. Scale bar: 50 µm. (B) Quantification of AAV2-hSyn-GFP transduction efficiency and specificity in RGCs. Efficiency is represented as the percentage of RBPMS-positive RGCs that also express GFP in the retinal flatmount. Specificity is represented as the percentage of GFP-positive cells that are also positive for RBPMS in the retinal section (n = 6 eyes from 4 mice). (C) Immunofluorescence staining of a retinal section and flatmount from C57 mice after 4 weeks of AAV9-GfaABC1D-eGFP infection. The colocalization of GFP (green) with GS (red) confirms Müller glia-specific expression. Scale bar: 50 µm. (D) Quantification of AAV9-GfaABC1D-GFP transduction efficiency and specificity in Müller glia. Efficiency is represented as the percentage of GS-positive Müller glia that also express GFP in the retinal flatmount. Specificity is represented as the percentage of GFP-positive cells that are also positive for GS in the retinal section (n = 6 eyes from 3 mice). Please click here to view a larger version of this figure.
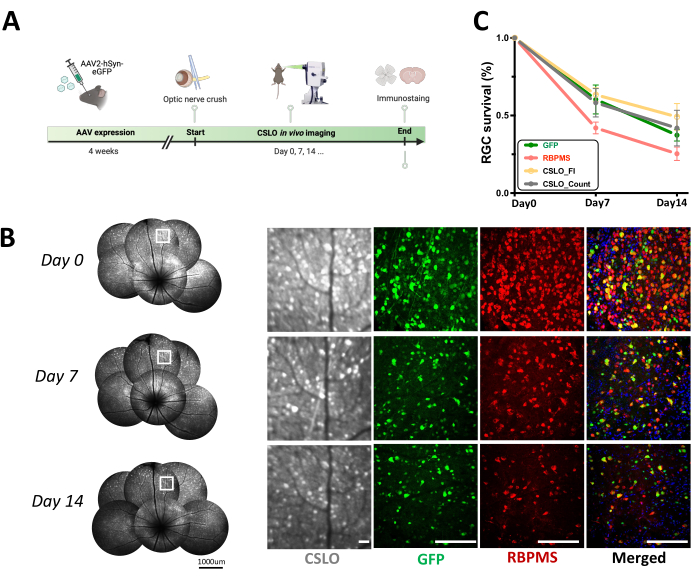
Figure 4: Longitudinal CSLO monitoring of RGC survival following acute optic nerve crush (ONC) in AAV2-hSyn-eGFP mice. (A) A schematic diagram of the experimental timeline of acute optic nerve injury after AAV infection. (B) Longitudinal CSLO imaging of the same retina in an AAV2-hSyn-eGFP mouse at baseline, 7 days, and 14 days post-ONC, visualizing RGCs expressing GFP signals. Magnified images of the boxed areas are shown on the right, alongside representative confocal microscopy images of retinal flatmounts demonstrating RGC morphology and distribution. Scale bars: 100 µm. (C) Quantification of RGC survival over two weeks post-ONC was assessed by measuring the percentage change in both cell number and CSLO fluorescent intensity relative to baseline (mean ± SEM). No significant differences were observed between groups at each time point (one-way ANOVA). Linear regression analysis revealed significant negative correlations between time post-injury and RGC survival metrics based on GFP fluorescence (r² = 0.945, p < 0.001), RBPMS staining (r² = 0.888, p < 0.001), CSLO-cell count (r² = 0.511, p < 0.001), and CSLO fluorescent intensity (r² = 0.7486, p < 0.001). n = 6 mice/group/time point. Please click here to view a larger version of this figure.

Figure 5: AAV-aided mapping of RGC axonal projections along the visual pathway. (A) Representative fluorescence microscopy images of a pair of optic nerves from an AAV2-hSyn-eGFP mouse, demonstrating GFP-labeled RGC axons (green) and colocalization with the anterograde axonal tracer cholera toxin B (CTB, magenta). The upper panel provides an overview of the entire optic nerves (scale bar: 1000 µm), while the lower panel offers magnified views of the boxed regions (scale bar: 100 µm). Two weeks post-ONC injury, both GFP and CTB signals show a marked reduction throughout the optic nerve (left side), indicating RGC axonal loss and disruption along the visual pathway. (B) RGC axonal projections to the superior colliculus (SC). Coronal brain sections from an AAV2-hSyn-eGFP mouse 2 weeks post-ONC highlight GFP-labeled RGC axons (green) and CTB staining (magenta) within the SC. Left (L): contralateral SC; Right (R): ipsilateral SC (ONC-injured side). Scale bar: 1000 µm. (C) A magnified view of the boxed region in (B) demonstrates the colocalization of GFP and CTB signals (merged image), highlighting RGC axonal bundles and their projection patterns within the SC. Scale bar: 100 µm. (D) RGC axonal projections to the lateral geniculate nucleus (LGN) following ONC, revealing GFP-labeled RGC axons (green) and CTB staining (magenta) within the LGN. Left (L): contralateral LGN; Right (R): ipsilateral LGN (ONC-injured side). Scale bar: 1000 µm. (E) A magnified view of the boxed region in (D) highlights RGC axonal bundles and their projection patterns within the LGN. Scale bar: 100 µm. Please click here to view a larger version of this figure.
Discussion
The presented protocol details a robust and accessible method for in vivo surveillance of specific retinal cell populations, harnessing the power of both AAV-mediated gene delivery and CSLO imaging. This approach offers several advantages over traditional methods, facilitating longitudinal studies of retinal cell dynamics and their responses to injury or disease under physiological or pathological conditions.
The success of this method hinges on several critical steps. Firstly, achieving optimal AAV transduction efficiency is pivotal for robust and specific expression of the fluorescent reporter in the target cell population. This can be influenced by factors such as AAV serotype selection, viral titer, and injection technique13,15,16. Secondly, proper animal handling and restraint during CSLO imaging are essential to minimize motion artifacts and maintain image quality. Employing a custom-built imaging platform and avoiding general anesthesia contribute to achieving clear and consistent visualization of retinal structures. Thirdly, careful optimization of CSLO imaging parameters, including focus, sensitivity, and averaging settings, is necessary to obtain high-resolution images with minimal noise and optimal signal-to-noise ratio26. Lastly, to address the potential limitations and ensure data reliability, we implemented stringent image quality control measures and increased animal numbers in our study design.
The Spectralis HRA CSLO system features automatic eye-tracking that enhances clinical longitudinal imaging. Its Automatic Real-Time (ART) tracking function can lock onto a specific retinal area during imaging, reducing motion artifacts.For human clinical imaging, the system also includes an algorithm for long-term tracking of the same anatomical retinal location27. However, these technologies are not optimized for rodent studies due to significant anatomical differences between human and rodent eyes. Rodent eyes are much smaller (3-4 mm diameter compared to 24 mm in humans) and lack certain landmarks like the macula, presenting unique challenges for automated tracking28. Consequently, researchers studying rodent models primarily rely on manual methods, using anatomical landmarks such as vessel patterns and the optic nerve head to locate specific retinal regions and maintain consistent imaging planes across sessions13,22. This approach is necessary not only due to limited landmarks in rodent eyes but also because of potential substantial retinal changes induced by experimental manipulations, which could confound automated systems not designed for these specific challenges. Thus, developing specialized eye-tracking and alignment algorithms for rodent retinal imaging represents a crucial future direction. Particularly, adapting long-term tracking algorithms for small animal use could significantly enhance the precision and reproducibility of longitudinal studies.
While traditional histological techniques require animal sacrifice and preclude longitudinal studies, this AAV-aided CSLO imaging method offers a non-invasive approach for monitoring retinal cell dynamics in vivo. This approach builds upon previous work demonstrating the utility of cSLO imaging for tracking longitudinal changes in specific retinal cells following injury, particularly in animal models utilizing transgenic fluorescent protein labeling19,21,22,29. These studies have highlighted the ability of CSLO to visualize a range of retinal cell events in vivo, including cell death, survival, and potentially even migration or process outgrowth. This protocol extends the application of this technique to study AAV-mediated transduction of specific retinal cell populations, allowing us to monitor the long-term effects of AAV delivery and transgene expression within the retina and providing crucial insights into the progression of retinal diseases and the efficacy of therapeutic interventions. Current clinical assessments using OCT primarily focus on RGC axon loss by measuring retinal nerve fiber layer thickness9, but they lack the resolution to assess individual RGC morphology or glial cell involvement. Although AO-SLO enables cellular-level imaging in the living human retina10, its complexity and limited availability restrict its use mainly in research settings. Additionally, differentiating glial cell types within AO-SLO often requires additional labeling methods30. This method overcomes these limitations by providing a more comprehensive and accessible approach for visualizing and tracking individual RGCs and other fluorescently labeled retinal cell populations over time. This opens avenues for investigating cellular responses under various conditions, aiding in the development of targeted therapies. Moreover, combining AAV-aided CSLO imaging with optogenetics31 or calcium imaging32 can further deepen the understanding of retinal cell function and its roles in visual processing, circuit development, and regeneration.
Despite the strength of this approach, some limitations and potential troubleshooting considerations should be acknowledged. Variability in AAV transduction efficiency can occur, necessitating optimization of injection parameters or viral titer for specific cell types or experimental conditions. CSLO imaging can be affected by factors such as corneal opacities, tear film instability, and eye movements, requiring careful monitoring and adjustments during image acquisition. Additionally, the limited penetration depth of CSLO restricts visualization to the inner retinal layers, potentially excluding deeper structures like photoreceptors33. The Spectralis cSLO system allows for clear visualization of distinct cellular features and patterns within the retina34. While this imaging modality can facilitate single-cell observation under certain conditions, as demonstrated in a previous work visualizing individual RGCs in sparsely labeled Thy1-YFP mice22, it can pose challenges for quantifying individual cells in densely labeled scenarios. For instance, in this study, the mass labeling of RGCs with eGFP following AAV-mediated retinal infection can sometimes hinder the clear visualization of individual cells, particularly in areas with variations in expression levels and cell overlap. This challenge is further compounded when imaging Müller glia, as their elongated morphology, spanning the entire retinal thickness35, makes it difficult to capture a single cell within a single focal plane using cSLO. The assessment of AAV9-mediated Müller glia infection efficiency, therefore, relies primarily on evaluating overall transduction patterns and distribution using CSLO imaging.
Furthermore, while ART averaging in the HRA system enhances image quality, the increased scan duration presents practical limitations, especially in awake animal imaging. The selected ART mean value of 15 is to balance image quality with scan time, providing sufficient noise reduction while maintaining a reasonable scan duration. To minimize animal discomfort and stress, imaging procedures need to be optimized for efficiency, employed trained personnel skilled in handling and imaging awake animals, and diligently monitored animals for signs of discomfort or corneal dryness throughout imaging sessions. These measures were crucial for ensuring both data quality and animal welfare, highlighting the importance of considering both factors when employing ART averaging for in vivo retinal imaging in awake animal models.
By acknowledging these limitations and potential sources of variability, researchers can take appropriate steps to optimize imaging protocols, minimize artifacts, and ensure the reliability and reproducibility of their data.
Overall, this accessible and readily implementable method offers a powerful means to investigate the complex interplay between RGCs and Müller glia, especially in response to injury or disease. By enabling researchers to visualize and quantify these dynamic interactions, this approach facilitates the discovery of novel cellular and molecular mechanisms underlying retinal pathologies. This, in turn, sheds light on the development of targeted therapeutic strategies and improved disease countermeasures.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (82130029). Figure 2A and Figure 4A were created with BioRender.com.
Materials
| 33 Gauge Needle | Hamilton Corp., Reno, NV, USA | 7803-05 | For intravitreal injection |
| 0.5% proparacaine | Santen Pharmaceutical Co., Ltd. | Topical Aneasthetics | |
| AAV2-hSyn-eGFP | OBiO Technology Corp., China | Virus titer: 2.7 x 1012 viral genomes (vg)/mL | |
| AAV9-GfaABC1D-eGFP | WZ Biosciences Inc., China | Virus titer: 4.5 x 1012 viral genomes (vg)/mL | |
| Betadine | Healthy medical company | 001651 | Topical Antiseptics |
| Corneal scelar forceps (toothed) | Mingren Eye Instruments, China | MR-F301A | For eyelid secure during intravitreal injection |
| Dumont 05# forceps | FST | 51-AGT5385 | For optic nerve crush |
| Graphpad prism | GraphPad Prism, USA | Graph drawing and statistical analysis | |
| HRA Spectralis | Heidelberg Engineering, GmbH, Dossenheim, Germany | "IR" and "FA" mode for CSLO imaging | |
| Image J/Fiji | National Institutes of Health, USA | Image processing | |
| Maxitrol antibiotic ointment | Alcon Laboratories, INC. USA | 0065-0631 | Topical antibiotics |
| Microliter Syringe | Hamilton Corp., Reno, NV, USA | 7633-01 | For intravitreal injection |
| Mydrin-P Ophthalmic solution | Santen Pharmaceutical Co.,Ltd, Japan | Pupil dilation | |
| Ophthalmic surgical microscope | Leica AG, Heerbrugg, Switzerland | M220 | For surgical operations |
| Pentorbarbitol Sodium | Sigma Aldrich, USA | 57-33-0 | Genereal Aneasthetics |
| Powerpoint | Microsoft Corporation, USA | Image alignment and cropping | |
| VISCOTEARS Liquid Gel (Carbomer) | Dr. Gerhard Mann, Chem.-Pharm. Fabrik, Germany | Topical lubricant |
References
- Cheung, C. Y., Ikram, M. K., Chen, C., Wong, T. Y. Imaging retina to study dementia and stroke. Prog Retin Eye Res. 57, 89-107 (2017).
- Ju, W. K., et al. Glaucomatous optic neuropathy: Mitochondrial dynamics, dysfunction and protection in retinal ganglion cells. Prog Retin Eye Res. 95, 101136 (2023).
- Vecino, E., Rodriguez, F. D., Ruzafa, N., Pereiro, X., Sharma, S. C. Glia-neuron interactions in the mammalian retina. Prog Retin Eye Res. 51, 1-40 (2016).
- Llorián-Salvador, M., Cabeza-Fernández, S., Gomez-Sanchez, J. A., De La Fuente, A. G. Glial cell alterations in diabetes-induced neurodegeneration. Cell Mol Life Sci. 81 (1), 47 (2024).
- Zhao, N., Hao, X. N., Huang, J. M., Song, Z. M., Tao, Y. Crosstalk between microglia and müller glia in the age-related macular degeneration: Role and therapeutic value of neuroinflammation. Aging Dis. 15 (3), 1132-1154 (2023).
- Minhas, G., Sharma, J., Khan, N. Cellular stress response and immune signaling in retinal ischemia-reperfusion injury. Front Immunol. 7, 444 (2016).
- Miao, Y., Zhao, G. L., Cheng, S., Wang, Z., Yang, X. L. Activation of retinal glial cells contributes to the degeneration of ganglion cells in experimental glaucoma. Prog Retin Eye Res. 93, 101169 (2023).
- Leung, C. K. S., et al. Diagnostic assessment of glaucoma and non-glaucomatous optic neuropathies via optical texture analysis of the retinal nerve fibre layer. Nat Biomed Eng. 6 (5), 593-604 (2022).
- Hood, D. C., et al. Detecting glaucoma with only OCT: Implications for the clinic, research, screening, and AI development. Prog Retin Eye Res. 90, 101052 (2022).
- Burns, S. A., Elsner, A. E., Sapoznik, K. A., Warner, R. L., Gast, T. J. Adaptive optics imaging of the human retina. Prog Retin Eye Res. 68, 1-30 (2019).
- Dinculescu, A., Glushakova, L., Min, S. H., Hauswirth, W. W. Adeno-associated virus-vectored gene therapy for retinal disease. Hum Gene Ther. 16 (6), 649-663 (2005).
- Nickells, R. W., Schmitt, H. M., Maes, M. E., Schlamp, C. L. AAV2-mediated transduction of the mouse retina after optic nerve injury. Invest Ophthalmol Vis Sci. 58 (14), 6091-6104 (2017).
- Cao, X., Yung, J., Mak, H., Leung, C. K. S. Factors governing the transduction efficiency of adeno-associated virus in the retinal ganglion cells following intravitreal injection. Gene Ther. 26 (3-4), 109-120 (2019).
- Vandenberghe, L. H., et al. Dosage thresholds for AAV2 and AAV8 photoreceptor gene therapy in monkey. Sci Transl Med. 3 (88), 88ra54 (2011).
- Gao, Y., et al. Develop an efficient and specific AAV-based labeling system for Muller glia in mice. Sci Rep. 12 (1), 22410 (2022).
- Nieuwenhuis, B., et al. Improving adeno-associated viral (AAV) vector-mediated transgene expression in retinal ganglion cells: Comparison of five promoters. Gene Ther. 30 (6), 503-519 (2023).
- Kawabata, H., et al. Improving cell-specific recombination using AAV vectors in the murine CNS by capsid and expression cassette optimization. Mol Ther Methods Clin Dev. 32 (1), 101185 (2024).
- Smith, C. A., Chauhan, B. C. In vivo imaging of adeno-associated viral vector labelled retinal ganglion cells. Sci Rep. 8 (1), 1490 (2018).
- Smith, C. A., Chauhan, B. C. Imaging retinal ganglion cells: Enabling experimental technology for clinical application. Prog Retin Eye Res. 44, 1-14 (2015).
- Leung, C. K. S., et al. Longitudinal profile of retinal ganglion cell damage after optic nerve crush with blue-light confocal scanning laser ophthalmoscopy. Invest Ophthalmol Vis Sci. 49 (11), 4898 (2008).
- Leung, C. K. S., et al. In vivo imaging of murine retinal ganglion cells. J Neurosci Methods. 168 (2), 475-478 (2008).
- Mak, H. K., Ng, S. H., Ren, T., Ye, C., Leung, C. K. S. Impact of PTEN/SOCS3 deletion on amelioration of dendritic shrinkage of retinal ganglion cells after optic nerve injury. Exp Eye Res. 192, 107938 (2020).
- Kokona, D., Jovanovic, J., Ebneter, A., Zinkernagel, M. S. In Vivo imaging of Cx3CR1GFP/GFP reporter mice with spectral-domain optical coherence tomography and scanning laser ophthalmoscopy. J Vis Exp. (129), e55984 (2017).
- Abbott, C. J., et al. Imaging axonal transport in the rat visual pathway. Biomed Opt Express. 4 (2), 364-386 (2013).
- Levkovitch-Verbin, H., et al. RGC death in mice after optic nerve crush injury: Oxidative stress and neuroprotection. Invest Ophthalmol Vis Sci. 41 (13), 4169-4174 (2000).
- LaRocca, F., et al. Optimization of confocal scanning laser ophthalmoscope design. J Biomed Opt. 18 (7), 076015 (2013).
- Vienola, K. V., et al. Real-time eye motion compensation for OCT imaging with tracking SLO. Biomed Opt Express. 3 (11), 2950-2963 (2012).
- Geng, Y., et al. Optical properties of the mouse eye. Biomed Opt Express. 2 (4), 717-738 (2011).
- Bosco, A., Romero, C. O., Ambati, B. K., Vetter, M. L. In vivo dynamics of retinal microglial activation during neurodegeneration: Confocal ophthalmoscopic imaging and cell morphometry in mouse glaucoma. J Vis Exp. (99), e52731 (2015).
- Joseph, A., Power, D., Schallek, J. Imaging the dynamics of individual processes of microglia in the living retina in vivo. Biomed Opt Express. 12 (10), 6157-6170 (2021).
- Chaffiol, A., et al. A new promoter allows optogenetic vision restoration with enhanced sensitivity in macaque retina. Mol Ther. 25 (11), 2546-2560 (2017).
- Li, L., et al. Longitudinal in vivo Ca2+ imaging reveals dynamic activity changes of diseased retinal ganglion cells at the single-cell level. Proc Natl Acad Sci U S A. 119 (48), e2206829119 (2022).
- Aumann, S., Donner, S., Fischer, J., Müller, F., Bille, J. F. Optical Coherence Tomography (OCT): Principle and technical realization. High-resolution imaging in microscopy and ophthalmology: New Frontiers in Biomedical Optics. , 59-85 (2019).
- Spaide, R. F., et al. Lateral resolution of a commercial optical coherence tomography instrument. Transl Vis Sci Technol. 11 (1), 28 (2022).
- Franze, K., et al. Muller cells are living optical fibers in the vertebrate retina. Proc Natl Acad Sci U S A. 104 (20), 8287-8292 (2007).
Tags

.
